Predicting: The future of health?
Assessing the potential, risks and appropriate role of AI-powered genomic health prediction in the UK health system
11 September 2024
Reading time: 231 minutes
Executive summary
Governments around the world are increasingly looking to emerging technologies to help deliver public services. This is especially the case in the UK, and in the context of healthcare. In recent years, hopes and predictions have proliferated about the potential of AI and genomics to transform the UK’s approach to medicine – with greater levels of efficiency, precision and personalisation held up as the prize for investment and adoption.
This report examines a technology at the vanguard of this promised transformation: AI-powered genomic health prediction (or AIGHP). AIGHP refers to a set of AI-driven techniques that enable predictions about people’s future health and drug responses to be made from genomic data.
AIGHP systems are powered by a form of genomic analysis known as polygenic scoring, which assesses the collective impact of multiple (individually small) genetic variations on the likelihood of a given person exhibiting a given trait (such as developing a particular disease), relative to the rest of the population. In recent years, AI systems have been applied to address the complexity and data intensity of some approaches to polygenic scoring. Though polygenic scoring can be conducted without AI, and while AIGHP is not yet the most common approach to polygenic scoring, some of the fastest developments in the field have come about as a result of AI, and this is expected to intensify.
While not yet widely used in healthcare, polygenic scoring has attracted considerable investment and expectation around the world. These trends, and the excitement behind them, are particularly marked in the UK, where research programmes such as Our Future Health (which proposes to use AI to accelerate inferences made from genomic analysis) have received considerable private and public investment, [1] [2] and where the UK Government[3] has published strategies citing the potential of AI-enhanced genomics to transform healthcare.[4] [5]
The project
This report is the second of two reports published as part of a two-year ‘futures’ research project conducted by the Ada Lovelace Institute in partnership with the Nuffield Council on Bioethics.[6] The project seeks to anticipate, assess and navigate the potential impacts of the convergence of AI and genomics over the coming five to ten years.
Our first report, DNA.I.,[7] investigated emerging trends at the intersection of AI and genomic science. Drawing on literature review, horizon scanning, scientometric analysis and extensive desk research, we identified AIGHP as one of the fastest and most clearly emerging and most potentially impactful capabilities in this space – and therefore as a capability requiring the attention of policymakers.
Building on this work, this report sets out the potential effects of AIGHP on the UK healthcare system in the near future. It also sets out a series of recommendations for policymakers focused on minimising the risks and maximising the benefits of this rapidly advancing capability. Our analysis is informed by a combination of our own desk-based research, expert interviews, an independently commissioned legal analysis, a series of scenario-mapping workshops with key experts and stakeholders, and a deliberative public engagement exercise. Our 10 recommendations are a synthesis of the insight generated from these sources, clarified by our analysis.
Our findings
AIGHP could bring significant benefits to healthcare
If appropriately integrated into a healthcare system, AIGHP could provide people with insight into their risk of developing particular diseases, inform beneficial lifestyle changes and help people be alert to symptoms of conditions for which they are at higher risk.
At a collective level, insight into variations in disease risk across the population could inform decisions about who to prioritise for screening and help with resource allocation by providing insight into groups or areas more likely to need particular treatments.
The use of AIGHP to improve understanding of how an individual might respond to a given drug or medication could allow for better prescribing practices, reduce waste, improve outcomes and avoid harmful side effects. If AIGHP enabled even marginal improvements, this could still be significant given the huge burden placed on the NHS by the ineffective use of drugs and widespread adverse drug reactions.[8]
To its champions, AIGHP also holds out the possibility of finally ushering in a prevention-focused approach to health and healthcare in the UK. We know that prevention is better than cure: better for people and far less costly to health services. We also know that earlier interventions have better outcomes. By providing people with better insight into their individual genomic health risks, AIGHP could enable people to better protect and promote their health, allowing them to stay healthier for longer, with far less reliance on expensive, curative interventions.
However, these benefits are not guaranteed. Large-scale deployment of AIGHP brings financial, ethical and service-level risks, and the science underlying these techniques is still being developed. The NHS will need to approach the deployment and cultivation of AIGHP deliberately and carefully if the benefits are to be realised.
The science around AIGHP remains uncertain
Despite the excitement surrounding AIGHP, there is substantial disagreement in the scientific community concerning the levels of accuracy and utility of such systems. Currently, polygenic scoring techniques (and by extension AIGHP) can suffer from poor accuracy levels when applied to certain individuals, and some polygenic scores are worse predictors of particular traits than more conventional diagnostic methods. Moreover, most current polygenic scoring systems are trained on datasets representing people with European genetic ancestry, meaning that they can perform badly for people of non-European genetic ancestry. Finally, for many common conditions, genomic variations appear to account for only a small proportion of disease risk. This limits the ability of polygenic scoring and AIGHP on their own to offer useful predictions about an individual’s health.
There is no consensus on whether these difficulties can be overcome, or in what time frame. Some argue that many of them will be resolved as datasets expand, become more diverse and improve in granularity, and as analytical techniques improve. Others maintain that the difficulties around polygenic scoring are more fundamental and cannot be resolved by improvements to scale, detail or sophistication.
AIGHP presents risks concerning privacy and discrimination
AIGHP is a technology that requires large amounts of sensitive personal data to operate, and which produces insight into people’s future characteristics that may otherwise be invisible. It therefore poses ethical questions concerning privacy, surveillance and novel forms of discrimination. AIGHP has several features that suggest its impact on privacy could be pronounced: it can be applied to a far wider proportion of the population than genetic tests, and it is difficult for a subject of AIGHP analysis to restrict the kinds of inferences that might be made about them from their data, now or in the future. Compared with other kinds of genomic research and AI predictive systems, AIGHP requires greater quantities of data, and typically more sensitive data. Genetic data can also produce information about people who the data subject is genetically linked to, intensifying some of the ethical challenges.
The use of AIGHP in healthcare must be sensitive to privacy concerns. In particular, policymakers will need to address models of AIGHP deployment in which the technology is a central part of care provision, which could make it practically difficult for people to opt out of sharing their genomic data. Reliance on AIGHP could sit uneasily with the principle that genomic data should be collected only with consent.
AIGHP also has the potential to both exacerbate existing forms and enable new forms of discrimination. This includes the risk of genomic discrimination, which occurs when a person or group is treated differently because they have genetic variations thought to be associated with a particular trait. In such cases, predictions about a person’s future health or likelihood of developing a particular disease could enable discrimination against people deemed to be more genetically predisposed to falling ill. A common worry, reflected in both the academic literature and the views of the UK public, is that people deemed more likely to fall ill because of their DNA might be offered worse terms for health insurance.
The conditions and ways in which AIGHP is deployed in the NHS will determine its success
Our research also highlights structural problems that could emerge from the way in which AIGHP is incorporated into health systems.
One potential issue is dependency, where the use of AIGHP leads to the NHS or its users losing control over the delivery or terms of healthcare provision. AIGHP deployment by the NHS is likely to require considerable amounts of data, compute and AI expertise. If the NHS looks to the private sector to provide these, it might have to navigate challenges around vendor lock-in, poor terms of access in the long run and the difficulty of auditing proprietary systems that will have a material impact on NHS decision making.
In addition to dependency, our research suggests that some approaches to using AIGHP in the NHS could reduce the resilience and effectiveness of the service. One risk is if the NHS channels money away from conventional, reactive care (or more conventional approaches to disease prevention) to the use of AIGHP systems, believing that such systems will reduce healthcare demand in the long run. If AIGHP systems prove unable to reduce demand to the degree expected, the NHS could find itself with a gap between unreduced demand and reduced supply of reactive services.
Getting the best out of AIGHP
The potential emergence of AIGHP systems presents an important, pressing and complex question for policymakers: how might AIGHP be integrated into our healthcare system in a way that maximises its potential benefits relative to the risks it poses and the resources it would require?
This report, which builds on close to two years of research, deliberation and expert engagement, is intended to help policymakers navigate this question. We bring together evidence on the science behind AIGHP, the benefits it could bring, the risks it could pose and the challenges of integrating novel technologies into the NHS. We then set out a series of concrete recommendations for UK policymakers, covering:
- the overall approach to and use of AIGHP that best suits the capabilities, limitations and requirements of the technology
- the conditions and protections that need to be in place before AIGHP can be responsibly deployed routinely in the NHS.
Our evidence suggests that while it has the potential to improve healthcare outcomes, AIGHP may currently be an ineffective tool for mass disease prevention and reducing healthcare demand at a population level. A wide deployment of AIGHP across the population could create greater exposure to the risks associated with the technology – and greater costs – in exchange for uncertain benefit.
Instead, the Government and NHS should move carefully and deliberately to enable the targeted use of AIGHP systems for cases in which there is a well-defined need for the insight they can provide. In the absence of good evidence of the viability of such a strategy, and a clear, credible plan to address the risks associated with it, the NHS should currently refrain from a wide rollout of AIGHP aimed at healthcare demand reduction. In this way, some of the most serious risks to patients and the public can be avoided without sacrificing AIGHP’s more certain benefits.
Both the NHS and the Government across England and the devolved nations must demonstrate that the following tests are met before AIGHP is rolled out for targeted uses:
- Minimum standards of accuracy and reliability for AIGHP systems should be defined, and mechanisms introduced for the ex ante and ongoing assessment of AIGHP system performance. Only systems that meet these standards should be licensed for use as medical devices in UK healthcare settings.
- The UK’s current legal and regulatory protections against surveillance and genomic discrimination need to be updated to ensure adequate protection against the risks that could be posed by the deployment of AIGHP systems. In particular:
- The UK’s data protection laws and consent practices need to be strengthened and clarified so they unambiguously protect the personal data required for AIGHP systems and provide members of the public with real control over if and how their data might be subjected to AIGHP.
- Protections against the use of AIGHP systems by insurers need to be strengthened, with a legal ban on the use of genomic data by UK insurers. Current arrangements, under which the UK insurance industry voluntarily pledges not to use genomic data to inform access to or terms of insurance products, are unlikely to reassure the public about how their genomic data might be used or to provide robust protection should AIGHP become more common.
We conclude with several recommendations for the Government and the NHS to ensure both a convincing scientific justification for the use of AIGHP and that appropriate safeguards are in place to make sure that deployments of AIGHP are well suited to its strengths and potential.
Recommendations
Recommendation 1: Any future reforms of UK data protection law should stipulate that genomic data should always be considered personal data. This would constitute an important revision to the current, context-dependent definition of personal data in UK data protection law (which holds that genomic data is only personal data when identifiable). Such changes should be designed to avoid circumstances in which determining whether a genomic dataset is personal data requires knowledge of the capabilities of particular data processors.
Recommendation 2: Any future reforms of UK data protection law should clarify how to interpret the UK General Data Protection Regulation (GDPR) definition of healthcare data. This should be done in a way that complements the current approach of the UK GDPR, under which healthcare data is defined by its ability to reveal information about a person’s health.
Specifically, the law should be clarified to:
- provide additional detail on what counts and does not count as revealing information about a person’s health status – and especially on what counts as revealing information about a person’s mental health status
- specify that only data capable of revealing information about a person’s health status on its own, or in combination with a limited number of other data points, should be considered healthcare data.
Following any such reform, the Information Commissioner’s Office should consider producing guidance setting out common examples of kinds of data that do and do not count as healthcare data.
Recommendation 3: Any future reforms of UK data protection law should clarify that biometric data should be considered special category data in all circumstances, regardless of the primary purposes for collection.
Recommendation 4: The Department for Health and Social Care, the General Medical Council and other relevant organisations should work together to create a more granular model of consent under which subjects can specify in greater detail what they want to be done with data they share.
This model should be used for patients sharing their genomic data for research or clinical purposes and for research participants. It should provide a new set of standardised options that are structured to enable people to explicitly opt out of particular uses of data, including sharing data with particular entities. Future reforms to UK data protection law should state clearly that these explicit vetoes mean that the ‘compatibility test’ (which requires entities seeking to process special category data for a new purpose to demonstrate compatibility with the original purpose for which consent was given) is not passed.
Recommendation 5: The Department for Health and Social Care and the General Medical Council should conduct a deliberative public engagement exercise to inform the development of the new, more granular model of consent proposed in recommendation 4.
Recommendation 6: Any future reforms to UK data protection law should strengthen, rather than weaken, protections around the repurposing of genomic and phenotype data for research purposes. Specifically, for genomic and phenotype data, any future amendments should preserve:
- the ‘transparency requirement’ around repurposing of special category data (so that entities processing special category genomic and phenotype personal data for a new purpose are still obliged to inform the data subject, even where the data is being processed for the purposes of research)
- the ‘compatibility test’ around repurposing of special category data (so that entities seeking to process special category genomic and phenotype personal data for a new purpose still have to demonstrate compatibility with the original purpose for which consent was given, even where the data is being processed for the purposes of research).
Recommendation 7:
The Government should develop an updated code of practice for the use of genetic and genomic data in the insurance industry. Building on the Association of British Insurers’ Code of Practice on Genetic Testing and Insurance, the new code should:
- prohibit the use of the results of predictive genetic and genomic tests for any kind of insurance, including life insurance, loss of earnings insurance and critical illness insurance
- explicitly define predictive genetic testing to include tests predicting both disease risk and drug responses, and to include testing looking at the risk of both genetic disease and common diseases
- define diagnostic genetic testing as applying to existing, symptomatic monogenic diseases, rather than common monogenic variants associated with disease risk; the latter should be explicitly considered predictive tests.
The Government should introduce primary legislation:
- requiring all insurers operating in the UK to comply with the updated code of practice
- enabling limited aspects of the code, such as monetary thresholds, to be amended by presenting the code before parliament, but making more substantive amendments to the code (including the kinds of genetic and genomics tests and insight an insurer may consider) impossible without the passage of new primary legislation.
Recommendation 8: The Government, the Equality and Human Rights Commission, relevant sector regulators and civil society should run a citizens’ assembly to explore the need for new primary legislation designed to address genomic discrimination, both in healthcare and in other domains, such as employment and education.
Recommendation 9: The Government, civil service and NHS should work to enable responsible, situational and high-impact deployments of AIGHP within the UK healthcare system. Such deployment should only be permitted where:
- adequate regulatory safeguards against surveillance and discrimination are introduced;
gaps in data protection and anti-discrimination law covered in this report and in the previous recommendations must be addressed in advance of any deployment of AIGHP systems in the NHS - the accuracy and reliability of AIGHP systems for different demographic groups reliably reaches a certain threshold; in its work on software and AI as a medical device, the Medicines and Healthcare Products Regulatory Agency should develop minimum standards of accuracy and efficacy for AIGHP systems and require any systems deployed in healthcare settings to meet them
- the NHS is demonstrably capable of and has committed to providing adequate and timely support for those who would be subject to AIGHP insight: any plans for deploying AIGHP in the NHS need to take account of the availability of genomic counselling for those subject to AIGHP insight; where the availability of genomic counselling is too low to provide it to everyone using AIGHP, and where there is no credible plan to expand access, AIGHP should not be deployed.
Where these conditions can be met, the Government and the NHS should work to enable the deployment of high-quality, carefully monitored AIGHP systems. To maximise impact, and to avoid cases where money and resources could deliver greater benefit elsewhere, AIGHP deployments should currently be restricted to cases in which there is a clear, clinically determined need for the extra insight provided by AIGHP, and where this benefit would outweigh any social or ethical risks, including discrimination and threats to privacy.
Recommendation 10: Given the risks and uncertainty about the accuracy and ability to reduce healthcare demand of AIGHP, the Department for Health and Social Care and the NHS should rule out the widespread deployment of AIGHP unless and until these uncertainties are resolved.
The Government, civil service and NHS should put in place safeguards to ensure that investments in uses of AIGHP are limited to those that are well evidenced, strategic and cost effective.
In funding, investment and resource allocation decision making and strategy, the NHS and Government should prioritise improving environmental determinants of healthcare outcomes over providing the whole population with insight into genomic variations in disease risk.
Any investments in AIGHP at scale for prevention should only be made where:
- this can be done in addition to, rather than in place of, addressing more fundamental problems with the health service
- there is clear evidence that providing of AIGHP to a large section of the population would result in significant and lasting reductions in demand for healthcare that could not be achieved more cost effectively through other interventions and investments
- concerns about privacy and individual control of genomic and healthcare data can be adequately addressed, and AIGHP can be rolled out so participation is optional rather than a de facto requirement of receiving adequate healthcare.
How to read this report
If you are a policymaker working on healthcare and technology:
- start with the chapter on the science and debate about AIGHP and its underlying technologies, which will provide you with an understanding of the opportunities and limits of these technologies for health prediction
- read through the four kinds of risks of these technologies to understand what kinds of novel issues AIGHP and the mass collection of genomic data can raise
- read our recommendations to understand what kinds of practical policy decisions the Government can make to create safeguards against these risks and ensure these technologies are adopted in a safe and effective manner.
If you work for the NHS:
- explore the chapter on the potential benefits of AIGHP to understand what value it might provide to the health service as a preventative tool, if the underlying science were to improve
- read through the four risks we identify for AIGHP technologies, especially the points on dependency and fragility that explore how the NHS should consider adopting and deploying these tools
- read our recommendations, particularly recommendations 9 and 10, which touch on how the NHS can adopt these technologies in a way that reduces risks and maximises their value.
If you are a regulator working on data protection or equalities:
- read through the four risks we identify for AIGHP technologies, especially the points on discrimination and privacy of genomics technologies, to understand what novel risks these technologies may raise
- read our recommendations, particularly recommendations 1 to 8, which touch on how existing equalities and data protection law can be updated to provide safeguards against the risks.
Introduction
Conversations about the appropriate role of technology in UK healthcare have been gaining momentum. The NHS ranked as the best healthcare system in the industrialised world as recently as 2017,[9] but it is now struggling with acute staff shortages,[10] backlogs[11] [12] and record low levels of satisfaction.[13] [14] At the same time, Government officials and technology companies claim that we are on the cusp of a technological revolution in medicine, with rapid advances in the life sciences, AI and data science poised to radically alter the way that countries keep their populations healthy and care for them when they fall ill.[15] [16] [17]
For the NHS and health policymakers, this poses an obvious, urgent and difficult question: how, if at all, might the technologies of this promised revolution be harnessed to deliver the NHS from its current problems? Can a crisis born of multiple causes, including sustained underfunding [18] [19] [20] [21] and rising demand, [22] be addressed by an emerging suite of medical and digital technologies focused on precision, prediction and automation? And if so, what would this look like?
This report interrogates these questions in the context of one of the emerging technologies often cited as having the potential to save the UK healthcare system: a suite of techniques we refer to as AI-powered genomic health prediction (AIGHP). These techniques apply AI-powered analysis to a person’s genomic data to make predictions about their health, their risk of developing non-transmissible diseases, and their response to drugs and medications.
In the UK, AIGHP has caught the attention of many working in health, business and politics, and is attracting considerable public and private investment. For example, £179 million in public and private funding has so far been invested in Our Future Health, a UK-Government-backed initiative whose work includes efforts to develop polygenic scores for common health conditions, ‘combining health and other data in conjunction with artificial intelligence’.[23] [24] The NHS has run smaller-scale pilots of genomic prediction to spot people at high risk of cardiovascular disease.[25] Over the last few years, the NHS and the Department of Health and Social Care (DHSC) have committed to investing in more forms of predictive and preventative treatments. This strategy has leaned into the promised power of AI and data-driven technologies to deliver better healthcare at lower cost.[26] [27]
There are good reasons for this interest. The kind of insight into disease risk and drug response variation that AIGHP systems could produce has several potential uses for healthcare systems and for individuals interested in better understanding and looking after their health. At an individual level, AIGHP’s ability to provide people with insight into their risk of developing a particular disease could inform beneficial lifestyle changes and help people to be more alert to symptoms of conditions for which they are at higher risk.
At a collective level, insight into variations in disease risk across the population could inform decisions about who to prioritise for disease screening and early treatment and could help with resource allocation decisions, by providing insight into groups or areas more likely to need particular treatments. AIGHP could also be used to improve understanding of how an individual might respond to a given drug or medication, which could allow for better prescribing practices, reducing waste, improving outcomes and avoiding harmful side effects. If AIGHP drove even marginal improvements in drug responses, this could still be significant given the huge burden placed on the NHS by the ineffective use of drugs and the numbers of adverse drug reactions.[28]
Compared with other countries, the UK is well placed to take advantage of emerging advances in genomic health prediction. It has a world-class genomics research sector,[29] longstanding strengths in the life sciences,[30] and a healthcare system that has already sought to integrate genomics-driven health and research initiatives such as the UK Biobank.[31] The UK is also one of the top nations in attracting AI companies and research talent, though its strengths compared with other countries are often overstated.[32] [33] [34]
The UK’s recent enthusiasm for AIGHP is also partly driven by hopes about what it might do for the NHS’s current challenges. AIGHP is cited (as set out in the box below) as a key enabler of a radically more preventative, resource-efficient health service – in which insights into people’s disease risks and potential responses to drugs empower them to avoid getting ill and enable them to be treated more efficiently and quickly when they do. This is an admirable ambition, but it is crucial to examine whether AIGHP will work in this way if integrated into the NHS, what risks the technology may raise, and if and how those risks can be overcome.
‘As healthcare costs continue to rise, investing in genomics-based screening … can help to mitigate disease through effective early intervention. We will shift away from a health and care system focused on diagnosing and treating illness and towards one that is based on preventing ill health and promoting wellbeing.’
The Office for Life Sciences and others, ‘Genome UK: The Futures of Healthcare’[35]
‘Everyone will have a genetic profile and doctors will use it to explain how you can potentially avoid getting those diseases you’re at risk of – or put you on a screening programme to catch the disease early.’
Dr Raghib Ali, ‘We Can Change the Whole Paradigm of Healthcare’[36]
For all the excitement about AIGHP, and the investment, there are pressing questions about whether it will be able to perform as promised; about the costs, opportunity costs and risks associated with deploying it widely; and about what it would mean to reconfigure the NHS’s current models of healthcare provision to take advantage of it.
This report examines these questions. We describe how AIGHP works and how it could be used in the UK healthcare system. We invite policymakers to consider the credibility and implications of the vision for developing AIGHP in the UK. We set out the necessary groundwork (in terms of regulations, law and policy) to enable responsible, routine deployments of AIGHP in the UK healthcare system. We also highlight models of AIGHP deployment that make best use of the technology’s capabilities, and that best negate its limitations and risks.
These considerations, and our findings, are especially topical as the Government meditates on its regulatory approach in several areas, with profound implications for if and how AIGHP is used. These include data protection reform, the regulation of medical devices, and the regulation of AI and automated decision-making. It is also topical in the wake of recent controversies around how medical data is stored, processed and shared, and current discussions about how the Government can assure the future of the NHS.
This report also offers precautionary advice for how the Government might deploy other emerging technologies in the delivery of public services.
Finally, while this report is focused on the application of a particular technology (AIGHP) to a particular sector (healthcare) in a particular place (the UK), many of the points raised have broader applicability. Many of the dynamics and concerns we describe around AIGHP in healthcare are similar to those seen with other AI‑ and data-driven systems in other parts of the public sector – including social care, benefits fraud detection, education and criminal justice. While this report is framed around the peculiarities of the NHS and the UK’s legal and regulatory system, many of the benefits, risks and ethical considerations posed by AIGHP remain relevant to other jurisdictions.
Project background and methodology
This report is the culmination of a two-year joint programme between the Ada Lovelace Institute and the Nuffield Council on Bioethics aimed at better understanding the potential societal and ethical implications of AI-enabled advances in genomic science.
The first half of this project focused on the most significant emerging trends at the intersection of AI and genomics. It identified AIGHP as a technology whose implications for healthcare needed further investigation. These findings and our analysis of them are detailed in our report DNA.I.: Early findings and emerging questions on the use of AI in genomics. Alongside extensive desk research and analysis, the first part of the project was informed by:
- A literature review conducted over spring and summer 2022 by independent researcher Dr Arianna Manzini and Tim Lee of the University of Edinburgh. This focused on how AI is being applied and is hoped to be applied to genomic science and on the current ethical, legal and policy debates around AI-powered genomics.
- A scientometric analysis carried out by the data science team at Nesta (the UK’s innovation agency) over summer and autumn 2022. The objective of this was to provide a data-driven understanding of trends in academic and industry research. It also aimed to offer insight into current and anticipated business models applying AI-powered genomics, and to identify the most significant public and private funders of research and development of these technologies – along with the biggest recipients of this investment.
- A horizon-scanning exercise, which used a form of the Delphi method to ask a panel of 13 external experts from academia, industry, medical science, Government and consultancies for their predictions about the most likely, impactful developments in AI-powered genomic science over the next five to ten years.
This report details the findings of the second half of the project, which explored the implications of the use of AIGHP in the UK healthcare system, and specifically in the NHS. In addition to extensive desk-based research and informal engagement with key experts and stakeholders, the major research analysis activities were:
- A scenario-mapping exercise, which used a technique known as morphological analysis to generate four distinct scenarios representing some of the ways that use of AIGHP in healthcare might develop and affect UK society over the next five to ten years, given different combinations of background conditions. The results did not directly inform the analysis and recommendations in this report but were used as stimulus material for our engagement with experts and the public.
- A deliberative public engagement exercise to understand public views and priorities on the governance, regulation and cultivation of AIGHP. This engaged 24 members of the English public, selected to be as close as possible to a representative sample of the total population. It sought their views on the possible worlds generated by the scenario-mapping exercise and asked them how they wanted the technology to be governed and regulated.
- A series of expert interviews and a policy development workshop with academics, policymakers, clinicians and civil servants. The interviews and workshop explored the risks posed by AIGHP systems in healthcare, the state of the current regulatory and governance environment, and the challenges and choices facing NHS policymakers regarding the implementation of AIGHP systems in the NHS.
This report has several limitations. First, it does not delve into the full range of ethical issues that AIGHP-based insights might raise, such as the potential to fundamentally change relationships between individuals, between clinicians and patients, and between the citizen and the state. Nor does it explore questions about the carbon footprint and environmental impact of computing at the scale required by AIGHP. While these topics are no less important than the ones raised here – and a broader public conversation is required about the deeper ethical questions posed by the emergence of genomic prediction technologies such as AIGHP – our focus is intentionally on the most immediate, tangible impacts of the technology and on issues with analogues to the uses of other AI-driven systems in the public sector.
Many of the deeper ethical questions absent from this document are signposted by the Nuffield Council on Bioethics’ recent work addressing the Government’s commitment to establish a ‘gold standard’ UK model for ethics across genomic healthcare and research. Further reading and resources are set out extensively in the report Towards a Gold Standard of Ethics across Genomic Healthcare and Research and its accompanying ‘resource bank’.[37] [38]
Finally, while our findings have broader geographical applicability, this report focuses primarily on how the issues presented by AIGHP might be managed by the NHS in England and the devolved nations of the United Kingdom. Most of our experts and all of our public engagement participants were from England, and we engaged in the greatest detail with NHS England’s policies.
Throughout this report, unless otherwise stated, ‘the Government’ refers to the UK Government in Westminster, as opposed to the governments of the devolved administrations. ‘The NHS’ refers to the four devolved health systems in the UK – NHS England, NHS Wales, NHS Scotland and NHS Northern Ireland.
The science and debate around AIGHP
Key scientific terms used in this report
Phenotype: The set of observable characteristics or traits of an organism, such as its physical appearance, behaviour and biological processes. Phenotypes result from the interaction of an organism’s genotype with its environment.
Phenotype data: By extension, this is information about the observable traits or characteristics of an organism.
Genotype: The specific genetic makeup of an organism, including all of its genes.
Genomic data: By extension, this is information about an organism’s complete set of DNA, including all of its genes.
Genetic data: Information about genes or parts of an organism’s DNA.
Polygenic trait / condition: A trait or condition that is influenced by multiple genes. Most common traits, such as height or risk of heart disease, are polygenic, as they result from the combined effects of many genes.
Monogenic trait / condition: A trait or condition that is determined by a single gene. Diseases such as cystic fibrosis and sickle cell disease are monogenic, as they are caused by mutations in one gene. Diseases caused by a single gene are sometimes referred to as Mendelian diseases or conditions.
What we mean by AI-powered genomic health prediction (AIGHP)
Throughout this report, ‘AIGHP’ refers to a set of AI and machine learning techniques that use genomic data to make predictions about:
- The probability of an individual developing common diseases and health conditions over their life course.
- How different individuals are likely to respond to medicines and treatments, given their DNA.
Figure 1: The three features of AI-powered genomic health prediction
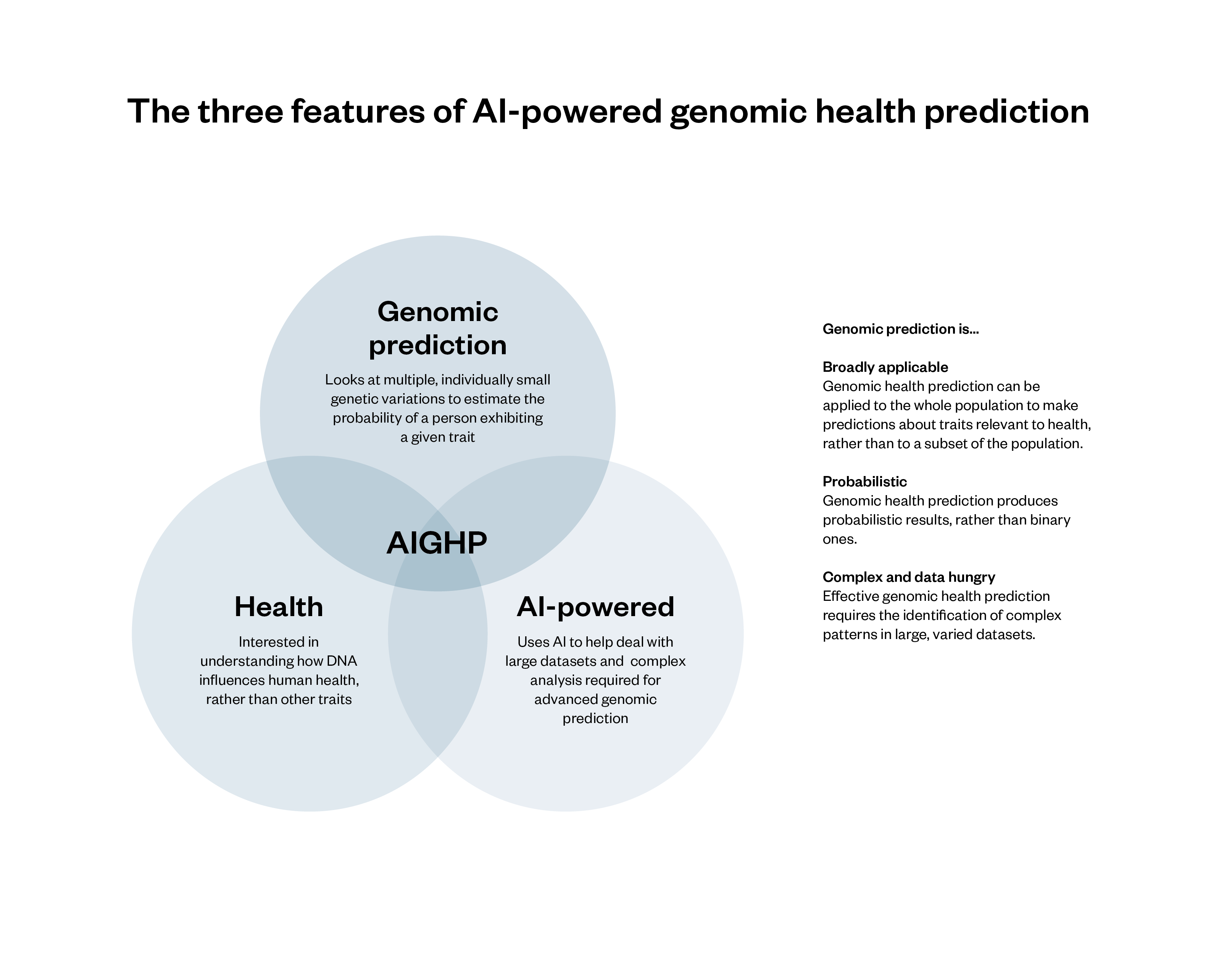
AI techniques can make traditional genomic health prediction methods faster, draw on more data and identify more inferences.
AIGHP has three broad features that distinguish it from other tools that use DNA to make inferences about people.
Health: AIGHP systems make predictions about traits relevant to health, such as a person’s likelihood of developing a disease or condition or their likely response to a drug or treatment.
Genomic prediction: AIGHP is an example of genomics (which looks at an organism’s complete set of genetic information) rather than genetics (which looks at the function and impact of specific genes). Specifically, AIGHP makes use of a form of genomic analysis known as polygenic scoring, which assesses the collective impact of multiple (individually small) genetic variants on the likelihood of a given person exhibiting a given trait, relative to the rest of the population.[39] The way that polygenic scoring works is set out below, but two features are especially important:
- Polygenic scoring is probabilistic. Polygenic scores do not definitively reveal whether someone has or will develop a given trait: they suggest whether a person has a higher or lower probability of having or developing a trait.[40] This is in contrast to genetic tests, which establish the presence of single gene variations associated with particular traits and produce binary results.[41]
- Polygenic scoring can be applied to a wider range of traits than genetic analysis, and can theoretically produce useful insight about a far larger section of the population. Genetic analysis and testing identify monogenic traits and diseases – those caused by a single genetic variation. In contrast, polygenic scoring deals with traits and diseases that are polygenic – where a person’s chances of developing them is influenced by multiple different genes. While genetic testing is useful for the 3.6 per cent of the UK population who have monogenic health conditions,[42] it provides limited insight for the other 96.4 per cent. By contrast, polygenic scoring can theoretically produce insight about everyone, regardless of whether they have a monogenic condition.Most diseases with a genetic component are not monogenic but polygenic, meaning that a given person’s chances of developing them will be determined by the interaction of multiple genes.[43] Polygenic scoring can therefore be used to predict the likelihood of a person developing some of the UK’s most common conditions, including cardiovascular disease,[44] diabetes[45] and some kinds of cancer.[46] (In such cases, polygenic scoring is often referred to as polygenic risk scoring, as it provides an indication of relative genomic disease risk.)
AI-powered: AIGHP systems make use of AI to help address the complexity and data intensity of some approaches to polygenic scoring. Polygenic scoring requires understanding and making sense of often extremely complex relationships between genomic data and the expression of traits. It has been significantly enhanced by the emergence of AI systems (especially machine learning and deep learning systems), which are very good at identifying and predicting patterns in large datasets.[47]
While genomic prediction and polygenic analysis can be conducted without the use of AI, some of the fastest developments in genomic prediction are the result of AI.[48] Moreover, as we detail below, AI may have a significant role in addressing some of the longstanding difficulties associated with polygenic scoring.
The current role of (and expectations for) AIGHP in the NHS
Since 2018, the NHS has used genomic science to improve diagnostics for people with rare diseases and to genotype different kinds of cancer treatment.[49] The use of AIGHP to make predictions about people’s future health for more common conditions is a relatively novel ambition. In its 2022 strategy Accelerating Genomic Medicine in the NHS, NHS England set out its plans to develop this functionality and discussed the benefits it could bring to healthcare.[50]
The use of AIGHP to predict the risk of a disease or responses to drugs is a laudable goal. If this approach works, it could help to create more personalised treatment plans for patients. Its use by the NHS therefore has the potential to reduce health service demand and improve patient outcomes. However, these benefits are uncertain.
The scientific basis for AIGHP is still unclear, and it may not provide useful, reliable or actionable insights for all disease risks or drug interventions.
How genomic health prediction (using polygenic scoring) works
Polygenic scoring comes from genome-wide association studies (GWAS). GWAS compare the DNA and observable traits of large groups of people to identify correlations between genetic variations – or combinations of variations – and phenotypic traits.
Polygenic scores for individuals are generated by analysing multiple sections of their DNA to establish the number and combination of genetic variants associated with a given trait. A person with a higher-than-average number of the genetic variants associated with the trait in question will be given a high score, indicating a higher probability that they have or will develop that trait, relative to the average person. A person with fewer of the gene variants will be given a lower polygenic score, indicating a lower probability of having or developing that trait.
Figure 2: The structure of typical genomic analysis and prediction
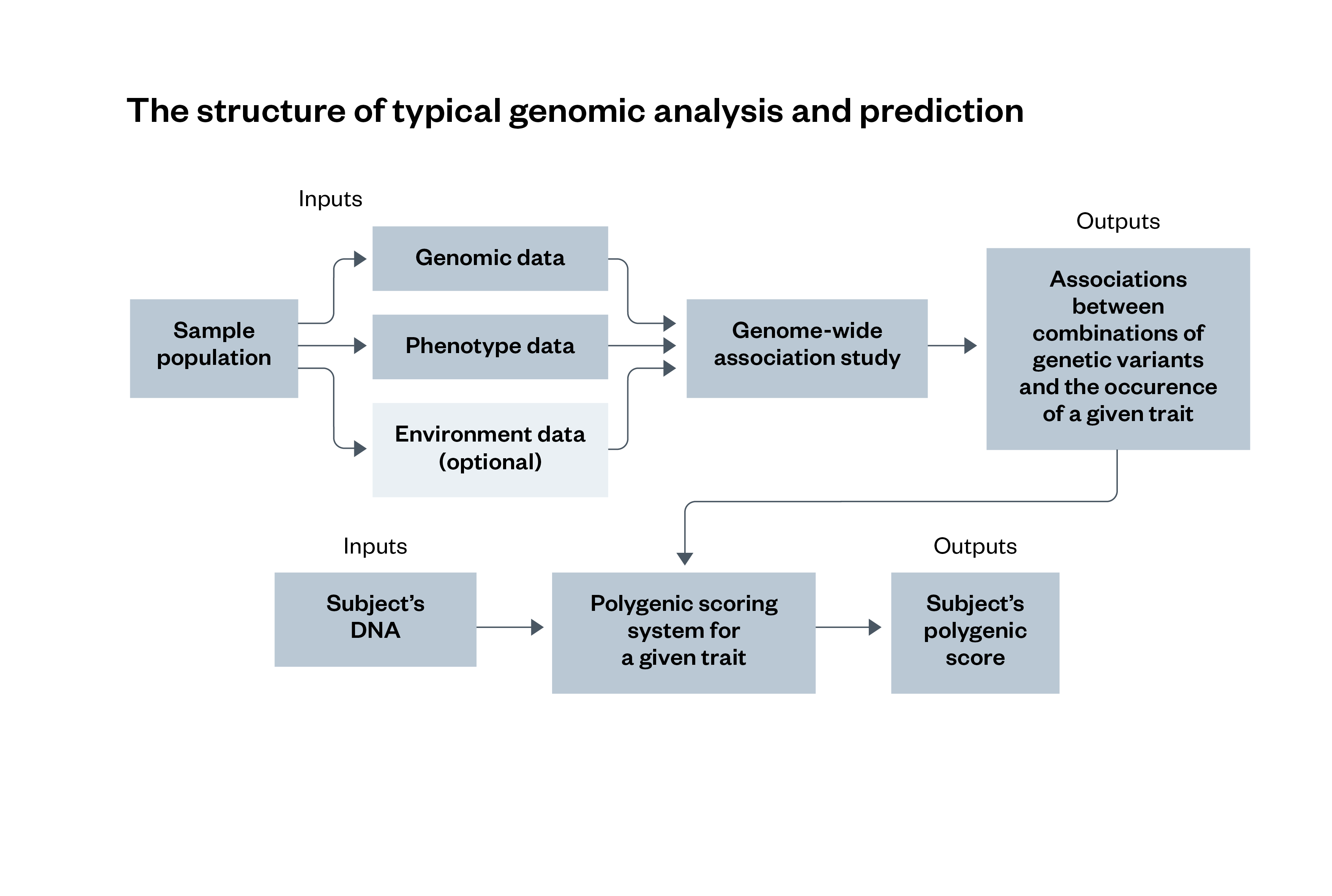
A GWAS involves a statistical analysis of three different types of data from a sample population – genomic data, phenotype data and environment data – that identify associations between different genetic variants and the occurrence of a specific trait. These associations are then compared with a subject’s DNA sample, which outputs a polygenic score for the traits they have. The use of environment data is optional for this process.
Figure 3: The use of AI-powered genomic health prediction techniques for genomic analysis and prediction
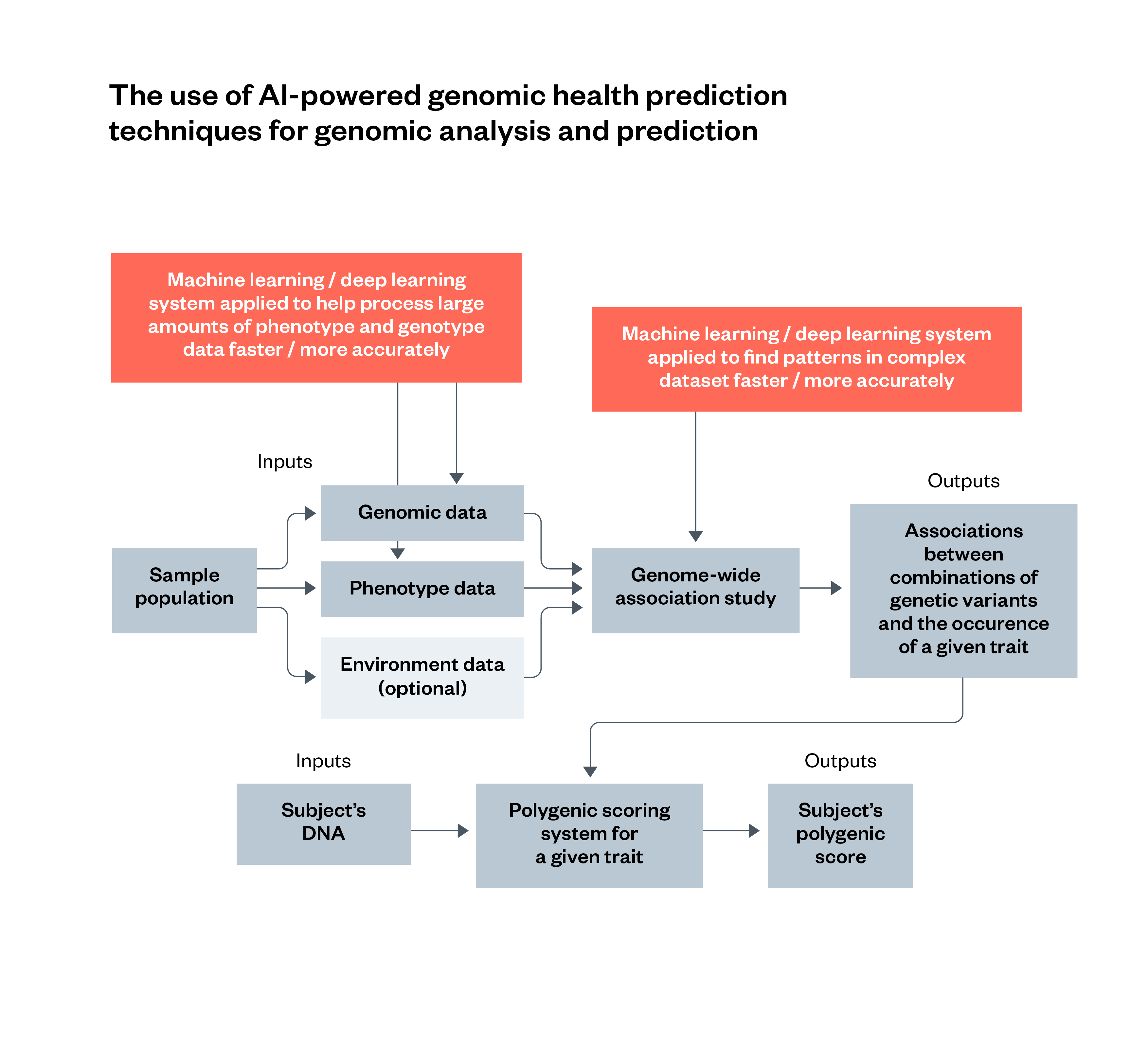 AIGHP involves the use of AI techniques such as machine learning or deep learning to enhance particular parts of a traditional genomic analysis. This includes analysing population-wide data, and finding patterns in the result of a GWAS. The use of environment data is optional for this process.
AIGHP involves the use of AI techniques such as machine learning or deep learning to enhance particular parts of a traditional genomic analysis. This includes analysing population-wide data, and finding patterns in the result of a GWAS. The use of environment data is optional for this process.
Challenges with genomic health prediction (using polygenic scoring)
Despite rapid developments in polygenic scoring, the accuracy, generalisability and utility of the technique is contested and is the subject of intense scientific debate.
The main challenges associated with current polygenic scoring techniques can be divided into three categories.
Poor absolute accuracy levels
Polygenic scoring systems are reasonably accurate at a population level, but they currently exhibit relatively low predictive accuracy for individuals.[51] [52] [53] For some traits, current polygenic scores are often worse predictors than more conventional diagnostic methods, such as blood tests or MRI scans.[54] Reasons for poor levels of polygenic scoring accuracy include:
- Small GWAS sample sizes (historical GWAS have been too small): One suggested reason for the poor accuracy of current polygenic scoring systems is that the sample sizes of historical GWAS have not been large enough to pick up the combined effects of genetic variations that individually have very small impacts on phenotype expression. It is hoped that as GWAS sample sizes increase, the accuracy of polygenic scoring systems will improve.[55]
- GWAS samples look at too few variants: A possible reason for the poor accuracy of polygenic scores is the limitations of the GWAS. Until recently, most GWAS were conducted using DNA microarrays, which look at variants at specific points in a person’s DNA sequence rather than at the entire sequence. Because of this, the complex interplay between different genetic variants (which can determine their impact on observable traits) may be invisible. However, recent advances have mitigated this issue. The number of variants assessed by GWAS has increased substantially in recent years, with modern GWAS looking at substantially more variants than earlier ones.[56] As whole-genome sequencing (which reads the entirety of a person’s DNA sequence) becomes cheaper and more practical,[57] it may well become the norm for future GWAS. This could lead to improvements in the accuracy of polygenic scoring systems.
- Poor data used for GWAS: As with other kinds of biomedical data, genomic and phenotype data are prone to noise and variation, so datasets often contain corrupted, incorrect or irrelevant data. Moreover, historical phenotype data, which is often used for GWAS, can be labelled in a way that reflects the biases of those labelling it (see box below).
Causes of ‘poor data’ in genomic science
Noise: One reason that noise in genomic data is difficult to address is that it is often created at the point of data collection or generation. This makes it harder to identify and account for later. In medical and healthcare contexts, noise is often a consequence of errors with equipment or techniques which lead to inaccurate results.[58] With genomic data, the DNA extraction process is often probabilistic and can therefore add erroneous data. Other reasons for noise include the loss of metadata, ambiguous criteria for applying particular categories or labels to data, or the inclusion of difficult edge cases that make accurate categorisation more challenging.
Bias in historical phenotype data: Historical phenotype data often reflects the prejudices of those responsible for the labelling.[59] Clinical notes recorded by physiatrists reflect the historical tendency to make different treatment recommendations for minority ethnic groups and female patients.[60]
- Confounding factors: This is perhaps one of the most fundamental problems for identifying the relationship between genomic variations and observable traits. Many non-genomic influences on observable traits (such as family and socioeconomic status) have a high degree of heritability and therefore often overlap with genomic variations. This can make it difficult to establish whether genomics, environment or some combination of the two is responsible for a given trait – and, in the latter case, the specific nature of the interaction between genomics and environment. Some argue that a move towards studies that specifically take into account the relationship between genes and environment could help to address the difficulties caused by confounding factors.
Poor portability of polygenic scoring across populations
One of the best-known problems with current polygenic scores is that most do not work well for people of non-European ancestry. This is because the findings of GWAS do not translate well to populations different from those on which they were trained,[61] [62] and to date, 83 per cent of GWAS have been conducted exclusively on cohorts of European genetic ancestry.[63]
There are various initiatives attempting to address poor diversity in genomic datasets for GWAS and to enable GWAS for people with non-European ancestry, including Our Future Health in the UK, the All of Us programme in the United States, and Human Heredity and Health in Africa (H3Africa).[64] However, recent research has argued that many of the datasets emerging from these projects have limitations that could limit the generalisability of research findings derived from them.[65]
Low predictive value compared with more conventional metrics
For many common conditions, genomic variations appear to account for a small proportion of disease risk. As a result, for most common diseases, more conventional and well-established risk factors such as smoking, obesity and socioeconomic deprivation may have a greater impact than a person’s DNA.[66]
While some of the possible developments mentioned above may show that genomics has a greater impact on disease risk for particular diseases, this remains an open question.
The apparently low impact of genomic variation on disease risk is not enough to make polygenic scoring useless. However, it does reduce the value of polygenic scores in informing health and clinical decisions at an individual level when used in isolation. In the case of common diseases, many argue that it is better to view polygenic scores as one contributor to a disease risk score that takes into account both genomic and non-genomic factors.
One way of doing this (which some polygenic scoring methods already incorporate) is to use genomic data to develop ‘combined risk scores’. These take a polygenic score and combine it with other data predictive of future health. The aim is to provide an incremental advance in predictive power over either a polygenic score looking purely at genomic variations or another scoring method used on its own. However, because many environmental and genetic factors are correlated, such approaches run the risk of double-counting risk factors.[67]
An alternative method for using genomics and other factors to predict the incidence of diseases and traits is to conduct studies that look at correlations between genomic variations and environmental factors, and observable traits. This is the approach taken by gene environment interaction studies, resulting in what are sometimes referred to as integrated risk scores. These kinds of studies are very complicated, however, and difficult to conduct on a large scale.
How AI could help to overcome problems with genomic prediction
AI is often described as a powerful tool to overcome the prohibitive complexity of genomic data and its interpretation. More specifically, AI (and especially machine learning and deep learning) is said to have the potential to help to address many of the difficulties with polygenic scoring mentioned above.
- AI could help to expand the size of GWAS: AI techniques are frequently cited as having the potential to help deal with the demands of increasingly large and complex GWAS. One of the biggest barriers to genomic analysis (and GWAS) is the availability of high-quality phenotype data. Machine learning techniques like natural language processing (NLP) – a computational technique aimed at analysing and synthesising natural language and speech – may provide a way to reduce the human resources required and speed up the preparation and interpretation of phenotype data (which is essential to GWAS).[68]
- AI could help to address issues with genomic data quality: Machine learning techniques can help to address issues with errors and noise in genomic data obtained through DNA sequencing.[69]
- AI could help to address difficulties with understanding the relationship between genomic and environmental factors: Perhaps the most fundamental issue for polygenic scoring is the difficulty of understanding how genomic and environmental factors (which are often highly correlated) act together to determine phenotype. Machine learning has been cited as a means of overcoming this problem.[70] For this reason, machine learning techniques, and especially deep neural networks, are increasingly being developed to generate polygenic scores in the hope that this will improve their accuracy and predictive ability. The use of deep neural networks may, for example, make it possible to understand the complex interplay between environmental and genomic risk factors for trait exhibition.[71] [72]
Figure 4: AI, machine learning and deep learning
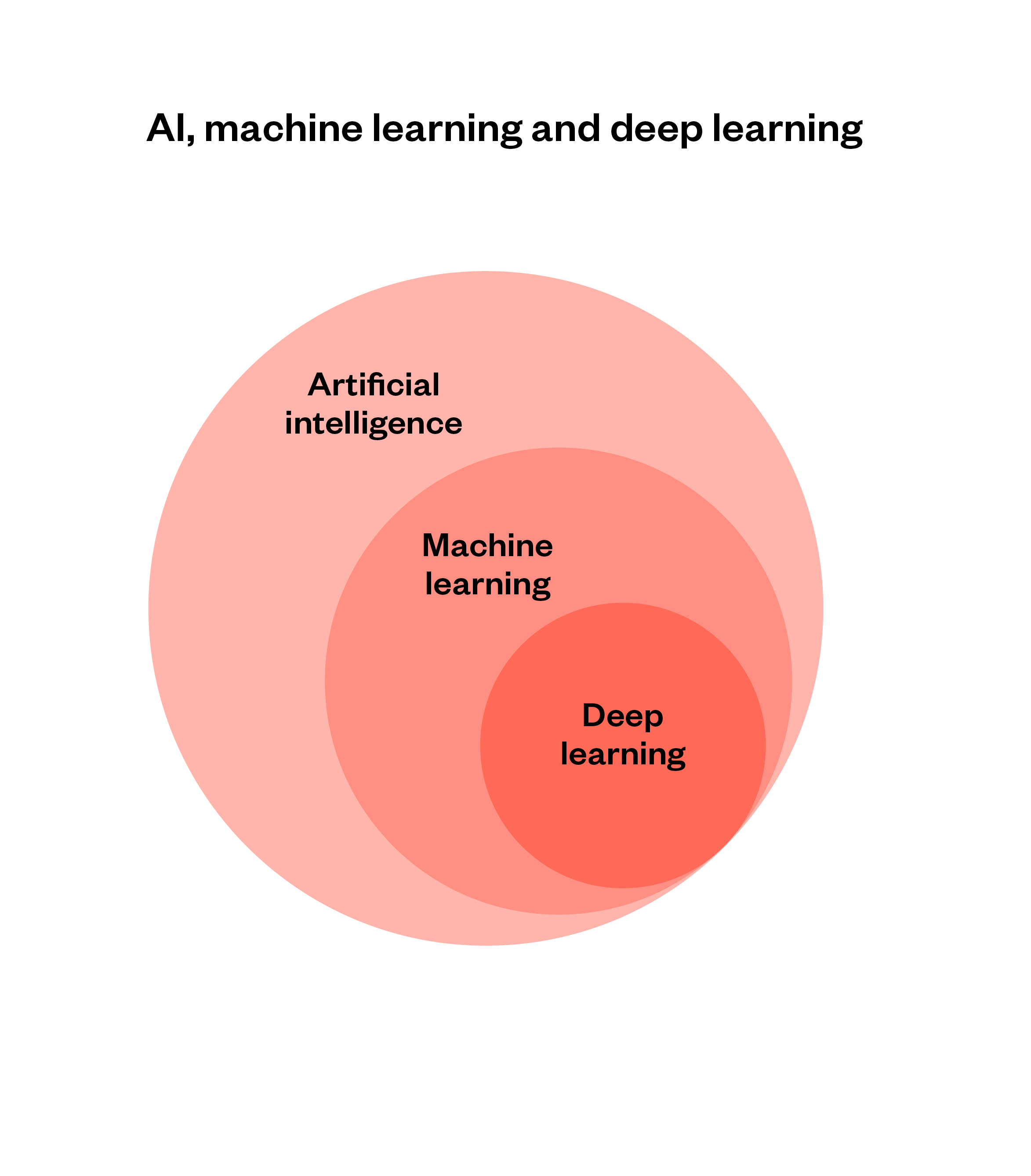
Machine learning is a branch of AI capable of recognising and predicting complex patterns in data, often with minimal assistance from humans.
Machine learning systems identify patterns in a dataset and formulate rules to predict the contents of new data points, which they can test and refine iteratively.
Deep learning is a subcategory of machine learning based on large artificial neural networks, also called deep neural networks. Neural networks are an approach to machine learning in which small computational units are connected in a way that is inspired by connections in the brain. Compared with traditional machine learning methods, deep learning systems have greater capacity to learn and process extremely large quantities of data.[73]
Potential benefits of AIGHP in healthcare
The fundamental scientific controversy regarding genomic prediction is about whether the difficulties with polygenic scoring described above can be overcome. Much of the optimism about the value of polygenic scoring assumes that as the quantity and quality of data increases, DNA sequencing techniques fall in cost, and the AI tools used to find complex associations in data become more sophisticated and powerful, the technique will become far more accurate and effective. Conversely, much of the scepticism surrounding polygenic scoring stems from the belief that these developments will not come about, regardless of progress in AI and elsewhere.
If the challenges of AIGHP are overcome, the deployment of AIGHP systems could improve healthcare provision. The two capabilities of AIGHP most commonly cited as having potential to improve healthcare are:
- AIGHP for drug response prediction: The use of AIGHP to improve understanding of how an individual might respond to a given drug or medication could allow for better prescribing, reduce waste, improve outcomes and avoid harmful side effects. This could be significant even if AIGHP enabled only marginal improvements, given the burden placed on the NHS by the ineffective use of drugs and widespread adverse drug reactions.[74]
- AIGHP for disease risk predictions: The use of AIGHP to provide insight into people’s genomic risk of developing common diseases could also be beneficial for healthcare and for population health.
AIGHP’s ability to produce insight into individuals’ risk of developing a particular disease could help to inform beneficial lifestyle changes, and could help people to be more alert to symptoms of conditions for which they are at higher risk.
AIGHP-generated insight into disease risk could also help healthcare interventions to be better targeted. In particular, the use of AIGHP to identify those at high risk of certain common diseases could allow the health service to offer them medications or other interventions to help lower their risk of developing the disease. AIGHP could also be used to inform decisions about who to screen for particular diseases. Those found to be at higher risk of a given condition could be prioritised for earlier or more frequent screening, and those at low or moderate risk could make the decision to have later and less frequent checks.
Some of the mechanisms by which AIGHP-driven disease risk prediction might improve healthcare and population health outcomes (as well as their limitations and associated trade-offs) are discussed in detail in subsequent sections of this report.
Risks posed by AIGHP in healthcare
Even if the scientific issues surrounding it are resolved, AIGHP could still pose significant risks to the people who might be subject to it – and, in some cases, to those with familial links to them. Some of these risks are functions of how the technology is deployed. Others may arise simply because of the availability of AIGHP and the insight it generates in health settings. Critically, these are harms that could arise even if AIGHP systems achieve high standards of accuracy and reliability and low levels of algorithmic bias – and which could challenge the benefits outlined in the above chapter.
Few of the risks we have identified are unique to AIGHP. Many are variations of well-established risks of AI systems, including data privacy, discrimination and the over-delegation of decision making to autonomous systems.
However, AIGHP can exacerbate these issues in surprising ways that may require new regulatory, design and integration solutions.
Our research identified four broad categories of potential harm from AIGHP:
- Surveillance (privacy): AIGHP both creates highly sensitive personal data and requires the collection and processing of more personal data than would otherwise be necessary for healthcare.
- Discrimination: The insights gained by using AIGHP could be used to discriminate between individuals and groups, e.g. on the basis of disease susceptibility.
- Dependency: The health system could become unable to administer healthcare effectively without the use of AIGHP, making it reliant on those providing the data, models and compute.
- Fragility: AIGHP could push healthcare into an excessively preventative mode of operation, at the expense of necessary reactive capacity.
The above categories were common themes in the findings of our commissioned literature review, which covered debates around the legal, ethical and social implications of AIGHP; in our deliberative public engagement exercise; and in our engagement with subject matter experts.
Some of the broadly held views of our deliberative public engagement participants are presented in boxes throughout this chapter.
Privacy and surveillance
Why privacy matters
Many of the concerns about AIGHP are ethical, relating to the privacy implications of the technology both for the data subject and, in some circumstances, for those with familial links to them. In addition to being a requirement of UK human rights and data protection law, privacy is also the cornerstone of trust in the relationship between patients and those who care for them.
Privacy is particularly important in the case of health and genomic data, and therefore in the use of AIGHP. According to many of our expert interview participants, it is the strongest safeguard against unwarranted profiling for disease or illness risks, the strongest protection against being judged and manipulated on the basis of perceived genomic predispositions, and the strongest safeguard against more formal genomic discrimination by institutions like insurers. Information about an individual’s genome also contains information about those who they are genetically linked to, creating further concerns about privacy. As we establish below, many of the other social and ethical harms that we identified around discrimination are intimately bound up with the threats posed to privacy by AIGHP.
Why AIGHP presents acute risks to privacy
As a technology whose existence and viability is premised on the creation, collection, processing and long-term storage of sensitive genomic, medical and other personal data, AIGHP presents significant challenges regarding privacy and data protection.
AIGHP is not the only use of genomic data or predictive analytics that has privacy implications. Insights from genomic diagnostic testing are notoriously sensitive, wrought with complex ethical challenges around a person’s – and their biological relatives’ – right to know, or not know, about a disease they may have.[75] Likewise, AI and predictive analytics systems in contexts such as social care or loan allocation typically require the collection of large amounts of personal data to produce personalised predictions.[76]
However, there are some features of AIGHP’s combination of AI prediction with genomic data that suggest its impact on privacy could be particularly pronounced.
- AIGHP has a wider reach than other genomic technologies. AIGHP can reveal information about a far larger section of the population than more established forms of genomic and genetic analysis such as diagnostic genetic testing. The latter is used to confirm or rule out the presence of genetic conditions and produces binary results. It is of great use in identifying and guiding the treatment of the 3.6 per cent of the UK estimated to have a genetic condition,[77] but it provides limited insight for the rest of the population.In contrast to genetic diagnostic testing, AIGHP is (theoretically) capable of being applied to everyone, producing insight about the relative disease risk and drug responses of those with and without genetic conditions. While this is a major potential advantage of genomic health prediction, it also raises the stakes regarding privacy, surveillance and discrimination.
- Inferences made by AIGHP are difficult for subjects to predict and control. AIGHP can allow a huge number of different inferences to be made about a person – and potentially those biologically linked to them – on the basis of their genomic data. In addition to susceptibility to common diseases and drug responses, traits such as risk-taking, substance abuse, intelligence and educational attainment may have genetic components and could theoretically be predicted using polygenic scoring techniques.[78]Given the rapid development of genomic prediction, it can be difficult for a person to understand when they share their data what inferences will be possible in the future. To share your genomic data is to share the key to an unknown and potentially vast amount of future insight about yourself.Moreover, since AIGHP will likely rely on whole-genome sequencing, participation in AIGHP will probably require a person to share their entire genetic code. This would make it practically difficult for them to limit what could be inferred about them in the future. Decisions about genomic data sharing for AIGHP are therefore likely to be all-or-nothing, with inherent uncertainty about how that data could be subsequently used to draw inferences about a person.
- AIGHP requires more sensitive data, creating a trade-off between privacy and accuracy. AIGHP systems require a wider array of sensitive data than other kinds of genomics research and other kinds of predictive analytic systems.[79] As mentioned earlier, accurate AIGHP systems will likely need to be developed through complex association studies that identify correlations between specific sets of genomic variations and environmental factors and particular phenotypic traits (such as variations in typical drug responses and disease risk). AIGHP systems therefore need to be trained on datasets that combine genomic, phenotype and demographic data.The need to combine so many datasets presents particular challenges for genomic privacy. First, it creates increased incentives for developers and deployers of AIGHP systems to collect and agglomerate large amounts of highly sensitive personal information about the population. Second, a common practice for respecting the privacy of individuals who have shared their medical and genomic data for research purposes is ensuring that they cannot be identified from the data provided. The problem with AIGHP is that the more demographic attributes are combined with phenotype and genomic information, the easier it becomes to reidentify data subjects. Research has shown that a relatively small number of data points can be used to reidentify research subjects.[80] [81] As a result, there may be a trade-off between the accuracy of an AIGHP system and the ability to ensure the privacy of those participating in research.
AIGHP and structural risks to privacy
The introduction of AIGHP into the UK healthcare system could also create structural pressures regarding the sharing and processing of personal data, including genomic data.
The centrality of AIGHP could make opting out difficult
Social pressures to share genomic data: If the use of insight generated by AIGHP becomes a fundamental component of healthcare, people could feel pressurised to share personal and genomic data.[82] Existing narratives around patients’ obligations, along with comments made by our deliberative public engagement participants,[83] suggest that the following views may become commonplace, prompting people to share more personal data than they might otherwise be comfortable with:
- Beneficiaries of AIGHP-guided care or public health schemes have an obligation to share their genomic and personal data to help develop, maintain and improve AIGHP systems. Anyone who fails to do so is ‘free-riding’ on the contributions of others.
- People interacting with health services have an obligation to share their genomic data to help the health service treat them as efficiently as possible. Anyone who refuses to share their genomic data is unnecessarily burdening the service.
These narratives could emerge where AIGHP is deployed in healthcare with the express intention of improving patient outcomes or preventing the emergence of illness. However, discussion with experts, prompted by our scenario-mapping exercise, suggests that they are most likely to emerge in the context of NHS-provided care, where views about the need to support health service provision appear to be more pronounced than in private healthcare.
These dynamics could also exacerbate existing health inequalities experienced by marginalised ethnic, minority or socio-demographic groups.
Experts noted that pressure to share data to help train AIGHP may be felt most acutely by members of groups for whom such systems are less accurate or less effective, such as those with non-European ancestry.
In the past, members of these groups have expressed serious concerns about sharing health data because the UK’s health and care system has historically misrepresented or mistreated them.[84]
Health-related pressures to share genomic data: Another source of pressure to share genomic data could be the fear of receiving worse care in a healthcare system that is configured to make use of AIGHP insight. Given the importance of healthcare quality to the public – and the concerns expressed by our deliberative public engagement participants around getting substandard treatment – many people might opt to share their genomic data for fear that failure to do so will result in poor care.
This fear could be justified, in a future in which AIGHP systems enable far more effective care than conventional approaches to medicine or in which health systems are no longer configured or equipped to provide effective healthcare without using AIGHP systems.
Financial pressures to share genomic data: A final potential source of pressure to share genomic data is the use of AIGHP insight by the health insurance industry. Academic literature, experience from other domains of insurance and our scenario-mapping analysis suggest that in a world in which private healthcare becomes more prominent in the UK and good-quality NHS care is less readily available, insurers’ use of AIGHP to inform access to and terms of coverage could create significant pressure for people to share their genomic data.
Insurers could make the disclosure of an individual’s genomic data (or the disease risk score generated by that data) a pre-condition of coverage. In a world in which NHS care could no longer be universally relied on, a failure to share genomic data would amount to a significant risk.
Alternatively, insurers could make access to the cheapest or best-value premiums contingent on disclosing genomic data (or disease risk insight). In such circumstances, for poorer individuals there would be significant financial pressure to share their genomic data.[85]
How reliance on AIGHP may raise challenges for the use of consent in healthcare
Another risk of AIGHP is that its widespread use could result in the health system moving away from or deprioritising consent as the primary mechanism for managing what can and cannot be done with a subject’s genomic data. This could have significant ethical implications for patients’ ability to control what happens to their data, encroach on privacy, and undermine trust in both therapeutic relationships and health systems.
Data protection law allows for multiple legitimate bases for the processing of sensitive personal data. Alongside explicit consent, Article 9 of the UK GDPR sets out nine other legitimate bases for processing special category data (such as genomic personal data).[86] According to some experts we spoke to, there is a chance that the increasing use of AIGHP systems could prompt a general move away from consent as the legal basis for health data processing, to some of the alternative legal bases.
Under current data protection law, researchers and healthcare professionals seeking to process genomic data must typically seek to acquire the explicit consent of the individual for each use of their data. Although highly protective of patient autonomy with respect to their data, these requirements make it challenging to use genomic data for iterative or open-ended research. The need to constantly reobtain consent to use the same dataset is a common complaint of many medical researchers. Such a complaint could become particularly commonplace in a world in which AIGHP systems are widely used and relied on.
In a future in which healthcare is dependent on AIGHP systems, the need to obtain consent for processing genomic data may be regarded as unsustainable. One concern that our expert interviews raised was whether consent is meaningful in the face of poor public understanding of the implications of sharing genomic data and pressures to do so: we cannot meaningfully consent to uses of our data that we do not understand. Other experts believed that the current requirement for explicit subject consent to process genomic data might set too high a bar for the kind of mass data collection and processing that the development, deployment and maintenance of AIGHP systems is likely to need.
Resolving the tension between patients’ control of their data and the data requirements of AIGHP systems poses serious ethical questions.
It also hinges on views about the ultimate utility of AIGHP‑ and data-driven approaches to healthcare more broadly.
Alternatively, dissatisfaction with consent requirements could emerge simply because too few people are volunteering to share their genomic (and other healthcare) data in the first place, either because of inertia or due to an active desire to withhold personal data.
There are strong reasons to regard a departure from consent as a threat to the privacy of anybody sharing their genomic data. Partly, this is due to questions about whether other bases for processing would prove sufficiently clear and restrictive to effectively guide the actions of data handlers and protect the interests of data subjects (a topic discussed in more detail in the following chapter). More fundamentally, however, consent is the only basis for data processing that does not rely on criteria determined by others, providing the data subject with a unique degree of control over if and when their data is processed.
Taken together with the worries articulated in the previous section, this points to a challenge for a consent-based model of genomic data protection. It may be difficult to find a model of consent that is both meaningful (in that it allows people to say no to their data being shared and processed) and sustainable (in that it is not an untenable barrier to genomic research and the use of genomic insight in healthcare). Failure to secure a meaningful balance could have serious implications for patient and public trust in the NHS.
The public’s views on privacy and surveillance
In our deliberative public engagement exercise, a common, strongly expressed sentiment was that genomic data, and associated healthcare data, is especially sensitive and deserves high levels of regulatory protection.
Participants felt that data subjects should be able to exercise a high degree of control over any data shared for AIGHP purposes:
- Any collection and processing of data for AIGHP needs to be conducted with the meaningful consent of subjects.
- Subjects should be able to retain agency over any data they have shared, and there should be transparency about how it is being used.
Discrimination
A recurring concern in our engagement with experts, our deliberative public engagement and the academic literature was that the insight generated by AIGHP systems could be used to discriminate between individuals – and in some cases groups of individuals – in a manner that would unfairly disadvantage some. This possibility was raised most frequently in relation to the risk of private health insurers discriminating against certain people with particular genomic traits. It also came up in the context of NHS care, where doctors or healthcare professionals might treat certain patients differently based on their genomic traits. Lastly, a concern raised by our experts and public engagement was the risk of interpersonal discrimination, where other people (if they learned the results of a person’s genomic test) might discriminate against or ostracise that person.
In the academic literature and in our deliberative public engagement, the possibility of genomic discrimination was a principal practical justification for the need for strict privacy and data protection standards for genomic data.
Genomic discrimination occurs when a person – or potentially a group of people – is treated differently on the basis of having genetic variations thought to be associated with a particular trait.[87]
While not all genomic discrimination is necessarily bad (prioritising people with high disease risk scores for treatment could be desirable), in the context of AIGHP the worry is that predictions about a person’s future health could enable discrimination against people deemed to be more genetically predisposed to falling ill (poor disease risks). For instance, people deemed more likely to fall ill because of their DNA might be offered worse terms of health insurance, or if that information were to be made available to an employer, they may also find it harder to get a job.
How AIGHP discrimination could interact with existing health and economic inequalities
There is extensive evidence of existing healthcare inequalities in the UK based on characteristics such as race, region and socioeconomic status.[88]
In contrast to these inequalities, the kind of discrimination enabled by AIGHP systems has the potential to be highly individualised. Rather than discriminating against a person for falling into a particular broad category (such as age, race or sex), AIGHP is likely to enable discrimination on the basis of traits unique to individuals (such as having a very specific combination of genetic variants associated with heightened disease risk).
AIGHP discrimination may not simply mirror the patterns of pre-existing forms of discrimination. Rather, genomic disease risk itself might give rise to discrimination, as well as interacting with or exacerbating existing discrimination.
However, even if people with good and bad genomic disease risks are randomly allocated across the population, the impacts of AIGHP discrimination are likely to be felt far more acutely by groups who are already disadvantaged.
While AIGHP could enable a relatively new form of discrimination, it could also compound existing inequalities in health outcomes. For example, the negative impact of being identified as having a higher risk of illness is likely to be far more pronounced for people from socioeconomically disadvantaged backgrounds if it results in higher health insurance premiums. Moreover, people from socioeconomically disadvantaged backgrounds may be less financially able than wealthier people of making behaviour and lifestyle modifications to compensate for having a higher risk of illness. In extreme cases, this could turn one of the potential advantages of AIGHP insight (the ability to take steps to counter genomic disease susceptibility) into an added source of stress, as people may feel they need to make changes to their lives that are financially difficult.
These considerations are particularly relevant in the UK, a country with some of the highest levels of economic inequality in Europe[89] and unequal health outcomes between different racial and socioeconomic groups. These health disparities were in full view during the COVID-19 pandemic, with those younger than 65 in the poorest 10 per cent of areas in England almost four times more likely to die from COVID-19 than those in the richest.[90] Likewise, in the first year of the pandemic, Black and South Asian people were twice as likely to die of COVID-19 as white people, when age and sex differences were taken into account.[91]
Discrimination and health insurance
In the academic literature, and in our engagement with experts and our deliberative public engagement exercise, health insurance markets were frequently cited as areas in which the availability of AIGHP insights could lead to harmful and more widespread discrimination. As entities whose business models depend on making accurate predictions of risk, health insurers might want to incorporate AIGHP-generated insight about individual and group disease risk into their decision-making processes.[92]
AIGHP could prove especially attractive to insurers for two reasons. First, unlike genetic testing – which produces useful insights only for the minority of the population with genetic conditions – AIGHP could yield insight about the future health of everyone. Second, because there is no necessary connection between a particular disease risk score and possession of any of the protected characteristics enshrined by UK equalities law, an insurer’s use of insight generated by AIGHP would be unlikely to count as unlawful discrimination. AIGHP could therefore enable health insurers to radically expand the use of factors out of individuals’ control to personalise the terms of coverage.
Specifically, health insurers might use AIGHP insights to inform:
- Who is and is not entitled to health insurance, with those with genomic risk scores that fail to meet a predetermined threshold ineligible for insurance.
- The price of insurance premiums, with those found to have worse genomic risk scores having to pay more.
- The terms of access to health insurance products. One possibility is that ‘shared-value insurance’ (where insurers offer favourable premiums or perks on the condition that the insured party makes lifestyle changes designed to minimise the chances of them making a claim)[93] [94] [95] could be combined with insurers’ use of AIGHP. Individuals with good genomic risk scores would be able to access premiums with few strings attached. By contrast, those with poor genomic risk scores might be able access health insurance, or pay favourable premiums, only if they demonstrate behaviours (e.g. health, diet or lifestyle habits or changes) that mitigate their poor genomic risk scores.
These practices could prove problematic for several reasons.
In our public engagement exercise, many participants remarked that some people having to pay more for insurance because of something they have no control over would be fundamentally unfair.
While participants acknowledged that health insurers already discriminate based on factors which people have no control over (such as the presence of pre-existing conditions), they found troubling the idea that this practice could become dramatically more widespread. Some participants also expressed disapproval of the idea that judgements about access to health insurance might be determined by probabilities, rather than by the presence or absence of pre-existing conditions, with predictions capable of being proven false.
This view appears to be widely shared, with several studies showing that the UK public[96] and publics from around the world take a dim view of the use of genomic data by insurers.[97] Recent incidents in Australia suggest that this opposition is one of the principal reasons that people do not want to share such data.[98]
Public worries seem to be echoed by many professionals and professional bodies. Almost three quarters of respondents to the UK Government’s recent consultation on the ABI (Association of British Insurers) Code of Practice on Genetic Testing and Insurance (the majority of whom were replying in a professional capacity or on behalf of organisations) expressed concerns about genetic testing and private health insurance, including worries that people would avoid genetic testing due to fears about results making insurance unaffordable.[99]
In addition to public dislike of the practice, one concern explored in the academic literature was that the use of genomically informed risk scores to determine access to, and the terms, of health insurance would undermine the ability of the insurance industry to pool risk across the population.
Currently, the inability of insurers to perfectly predict the risk posed by individuals leads to premiums being based on averages, with some people who present a higher risk paying less and some people at a lower risk paying more than they should (in strict actuarial terms). There is a debate about whether this constitutes an inefficiency and unfairness to be rightly overcome through improvements to risk prediction, or whether it allows insurance to be an important vehicle for social solidarity.[100] [101]
On the latter view, insurance can be a valuable mechanism by which those at low risk subsidise those at higher risk, enabling risks that are unevenly distributed across the population to be borne more equally.[102] [103]
A variation of this objection to insurers’ use of AIGHP insight is that while actuarially fair price setting might result in cheaper premiums for those deemed to be low genomic insurance risks, it would also result in health insurance becoming more expensive for those deemed to be high insurance risks: precisely the people most in need of that insurance.[104] In other words, the use of AIGHP to personalise insurance premiums could result in it becoming more difficult for those most in need of health insurance to acquire it affordably. By extension, attempts to use AIGHP insight to offer more favourable rates to those with high genomic risks through shared-value insurance[105] could also result in unequal outcomes.
Experts we engaged with commented on the possibility of AIGHP insight intensifying the ‘responsibilisation’ of healthcare, with patients increasingly expected to identify and manage their own disease risk.
This worry about ‘responsibilisation’ is echoed in the academic literature. While expectations regarding patients’ behaviour are nothing new, research has argued that – by blurring the boundary between health-related behaviours and broader lifestyle choices – the use of data-based predictive tools could lead to a broadening out of what is expected of patients if they are to be considered deserving of care.[106] With the rollout of predictive tools in healthcare, ‘Taking care of one’s health, and being a good patient, may become a full-time duty even for those who consider themselves to be healthy’.[107]
Those with poor disease risk scores could be subject to far more, and more onerous, requirements to maintain access to insurance coverage than those who are lower-risk. As remarked on by participants in our deliberative public engagement exercise, an individual’s capacity to adhere to lifestyle changes is often contingent on factors beyond their immediate control. For those with bad risk scores, genomic shared-value insurance could therefore attach significant hurdles to accessing affordable health coverage.
Discrimination in the NHS and the public sector
Many of the forms of discrimination discussed above in the context of private health insurance are likely to have analogues in public sector health provision.
Existing debates around who is deserving and undeserving of NHS care – and in particular, debates about whether there should be limits to the amount of NHS care provided to people seen to be suffering from self-imposed conditions[108] [109] – could well take on a new dimension with the AIGHP insight into individual disease risk.
The availability of individual disease risk scores could fuel the idea that (1) people – and particularly people with high genomic disease risks – have a responsibility to prevent themselves from getting ill in the first place and (2) those who get ill despite being equipped with insight into their disease risk – having failed to act on this knowledge – are less entitled to taxpayer-funded healthcare.
As with the use of genomic shared-value insurance discussed above, the emergence of these narratives could place extreme, unfair pressure on those with high disease risk scores to avoid burdening the NHS. Likewise, the ability to comply with health advice could be unevenly distributed.
The public’s views on discrimination
One of the most common concerns expressed by participants was the use of AIGHP by insurers. The prospect of insurers using genomic health predictions to determine insurance premiums was generally thought to be unfair, to place undue stress and burden on those affected, and to risk compounding existing health inequalities.
Participants also voiced concerns about the potential for AIGHP to be used for other discriminatory purposes, such as enabling interpersonal discrimination and discrimination by employers.
Dependency
The third category of risks associated with AIGHP is dependency. This refers to scenarios in which different entities, groups and people experience an undesirable loss of control over the delivery or the terms of their healthcare because of the use of AIGHP systems. Specifically, it covers ways that AIGHP systems could disempower the NHS, clinicians and patients.
NHS dependency on AIGHP systems and those providing them
The deployment of AIGHP by the NHS is likely to require considerable amounts of data, compute and AI expertise. While it would be possible for the NHS to develop this capacity with sufficient public investment, current approaches to NHS investment and commissioning tend towards the participation of the private sector. There are particular risks associated with relying on an external provider for a system such as AIGHP, which:
- could likely only be provided by one of a handful of extremely large technology companies, whose business models are based around the accumulation of large datasets and the monetisation of insight derived from that data
- would likely be hard to replace, or transition away from once integrated into NHS processes
- would form a key input, and sometimes a determining factor, in NHS decision making at a micro level (for instance, informing individual clinical decisions) and possibly also at a macro level (for instance, informing broader commissioning and resource allocation decisions).
Perhaps the biggest risk to the NHS is vendor lock-in. As AIGHP systems become integrated into NHS workflows, and other services and processes get designed around them, the NHS could struggle to cope without access to such systems. Unable to function without AIGHP services and with few, if any, other entities able to provide these services, the NHS may have to accept any updated terms offered by its current provider. This lock-in, whereby the high costs of switching and lack of alternative providers render a customer dependent on a particular provider regardless of whether it provides value or works for the customer,[110] is common in healthcare contexts[111] [112] and represents a concern in many NHS procurement debates.[113]
A more specific variation of this risk is that the NHS may find itself pressured to share patient data with an AIGHP provider in exchange for access (or discounted access) to AIGHP systems. For example, the Royal Free Hospital notably shared patient data with Google DeepMind for just £1 in 2014.[114] More recently, Tony Blair and William Hague have advocated that the NHS sell its patient data to fund medical advances.[115]
Despite calls for large-scale sharing of patient data with commercial organisations to drive improvements in the NHS, there are significant risks. Most fundamentally, an arrangement of this nature would challenge the ability of the NHS to respect patient privacy and principles of purpose limitation (enshrined in the UK GDPR) around the sharing of personal and special category data. The data required for the training of AIGHP systems may be difficult to anonymise. More details on the UK’s data protection rules and consent practices are provided at the beginning of the next chapter.
Such an arrangement could damage public trust in the NHS and its use of AIGHP. Polling and public engagement exercises (including our own deliberative public engagement) have consistently shown that the public are happy to share their data with the NHS but hostile to sharing it with private companies.[116] [117] Recent history shows strong and enduring public and professional concerns about health data privacy. Dislike (and willingness to opt out) of schemes perceived to violate privacy is borne out by the backlash to care.data,[118] the Royal Free’s data-sharing deal with DeepMind,[119] [120] the General Practice Data for Planning and Research (GPDPR) programme,[121] and the Palantir bid to provide the NHS data-sharing infrastructure.[122]
Even if patient data could be effectively and reliably anonymised, there would still be difficult questions about the desirability of a private company using health data collected by a taxpayer-funded public service to develop and train proprietary AI systems and models.
A final risk presented by AIGHP systems provided by third parties is if the NHS were unable to understand or audit their decision-making processes. A commonly voiced concern about AI is the opacity of its decision-making processes and that the underlying reasoning behind such decision making can be difficult for a human to interrogate or understand.[123] [124] This problem could be compounded if private providers of AIGHP systems treat them as proprietary and do not allow the NHS the necessary access to the system – in particular, its weights and parameters – so that its performance can be audited. Previous examples of the NHS sharing data in exchange for insight, such as the Royal Free / DeepMind collaboration, demonstrate that AI systems developed using NHS data can remain the intellectual property of the private collaborator even if the NHS gets access to the insight generated.
An arrangement of this kind could undermine the ability of the NHS and individual clinicians to understand the basis for their decision making. Risk calculations and predictions are not wholly objective: the way their parameters are set reflects other values and trade-offs. It is entirely possible that two different proprietary AIGHP systems trained on the same data would return meaningfully different predictions.
Clinician and patient dependency and disempowerment
In addition to the risk of a structural transfer of power away from the NHS and towards the providers of AIGHP systems, the use of AIGHP could also disempower individual clinicians and patients.
As well as informing prevention, insight generated by AIGHP systems could inform clinical decision making, with disease risk scoring and pharmacogenomics feeding into diagnostic and treatment decisions. The incorporation of AIGHP systems into clinical workflows could contribute to a broader set of problems emerging from increased reliance on AI-powered predictive and clinical decision support systems.
First, the academic literature, along with our deliberative public engagement and engagement with experts, revealed concerns that clinicians could come to over-rely on the judgements on AI systems (including AIGHP). In high-stakes settings and under significant time pressure, clinicians could well succumb to automation bias, lacking the capacity to scrutinise the outputs of AI systems and finding it easier (and professionally less risky) to defer to them rather than risk having to defend deviating from their recommendations. Over the long term, this tendency to defer to AI systems could become entrenched, leading to the deskilling of the workforce.[125]
One potential knock-on effect of reliance on AIGHP and clinical decision support systems, mentioned in the academic literature and in our deliberative public engagement, is a narrowing of the scope of the evidence that the medical system can routinely consider. Whereas human medical professionals can supplement and contextualise standardised medical data with less standardised informational inputs (such as information provided by patients), the variety and format of information that AI systems can use is more limited.[126]
A common complaint about making medical data legible to analytics and AI systems is that it involves scrubbing out much of the complexity that makes it meaningful. Some academics have suggested that deference to clinical decision support systems could undermine the ability of medical professionals to incorporate non-machine-readable information into clinical decision making.[127]
The inability of medical systems to fully take account of idiosyncratic, non-standard and contextual health information could also have an adverse effect on the relationship between clinicians and patients. A critical part of the clinician–patient relationship is the opportunity for patients to communicate their experiences, needs and preferences, and the ability of the clinician to take this information into account. If clinicians routinely defer to AI-powered clinical decision support systems, their ability to account for patients’ professed needs and preferences, and patients’ ability to be heard, could be undermined.[128] Although the extent of future use of AIGHP is uncertain, these scenarios highlight the possibility of AIGHP, if it is imperfectly used, to alter the nature of therapeutic relationships and to undermine trust in both medical decision making and health systems.
The public’s views on dependency
Many participants expressed worries about health systems coming to over-rely on AIGHP systems to support clinical decision making. Concerns revolved around the potential for AIGHP systems to make mistakes, as well as the more structural risk that their availability could lead health systems to train and hire fewer staff. It was noted that this could make health systems especially vulnerable in the event of a failure of AIGHP systems.
Participants were generally more comfortable with the NHS having genomic and phenotype data than the private sector. Indeed, many of the concerns around data processing and storage concerned this data leaving the NHS and finding its way into the hands of third parties, particularly the private sector.
Fragility
A final risk identified by our research is that the widespread adoption of AIGHP could reduce the health system’s ability to cope with pressures or sudden fluctuations in demand. There are three broad means by which AIGHP systems might render the health system more fragile, which we consider in turn.
AIGHP leads to an uneven division of labour between the NHS and the private sector
One risk raised both by the experts we engaged with and the participants in our deliberative public engagement exercise was the potential for AIGHP to exacerbate and hasten the emergence of an undesirable division of labour between the NHS and private healthcare providers.
Both experts and the public voiced the concern that in a near future in which private healthcare forms a more prominent part of the UK’s healthcare mix,[129] [130] [131] the ability of private health insurers to use AIGHP insight to inform the premiums paid by different people could lead to:
- those with higher genomic disease risk being priced out of private health insurance and having to use NHS services instead
- those with lower genomic disease risk being enticed (by cheaper premiums) into the private health sector, where they may have otherwise used NHS services.
As a consequence, the average user of NHS services could end up being a worse disease risk (and therefore requiring more care) than in:
- the status quo, where the NHS cares for the vast majority of the population
- circumstances in which private healthcare is more prominent but those with lower and higher genomic disease risk are more evenly distributed between the NHS and the private sector.
Unless NHS funding were adjusted upwards to address this potential change, the uneven distribution of disease risk between the NHS and private healthcare could put substantial strain on the NHS.
Appeal to the potential of AIGHP diverts attention away from urgent debates about the adequacy of current NHS funding
Genomic medicine (and especially the capability of disease risk scoring that we capture under the term ‘AIGHP’) is increasingly cited as one of a suite of technological advances with the capacity to save the NHS from the challenges facing it.
A prominent idea (discussed in more detail in the final chapter of this report) is that AIGHP systems will allow the NHS to transition to a more preventative footing, thereby radically reducing demand for healthcare services.
Regardless of whether it is introduced or used in this way, the ability of politicians and policymakers to cite AIGHP (and other emerging technologies) as a future source of improved NHS efficiency could reduce the pressure to address more immediate questions about NHS funding and public health.
AIGHP prevention is implemented but fails to produce the reduction in demand promised
Another risk discussed with the experts we engaged with concerned the use of AIGHP as a means to improve public health and prevention measures – and in so doing, reduce healthcare demand.
The idea that insight into disease risk generated by AIGHP could enable a far more targeted approach to disease prevention and public health is common in the health policy world. Specifically, by providing improved insight into people’s disease risk, it is argued, AIGHP could potentially help some people to avoid certain health problems – and could catch developing illnesses earlier.
There is, however, ongoing debate about how effective AIGHP could be at reducing overall healthcare demand, especially compared with more conventional interventions.[132] [133] [134]
Given these uncertainties, policymakers will need to be careful when considering investment in AIGHP as a public health tool. This will be particularly important in cases where investment in AIGHP might crowd out:
- investment in more effective tried-and-tested approaches to disease prevention and public health
- investment in reactive services which respond once disease is already symptomatic; this could lead to a gap between capacity to deal with acute and chronic illness and unreduced demand, thereby making the service less resilient.
Uncertainties around AIGHP bringing about substantial reductions in healthcare demand
Three of the mechanisms by which the use of AIGHP could potentially reduce healthcare demand are:
- Targeted intervention: AIGHP insight could help the NHS to provide early interventions and medications for those at elevated risk of common diseases.
- Targeted screening: AIGHP insight could help the NHS to target screening programmes towards those at elevated risk of common diseases.
- Lifestyle change: AIGHP insight about disease risk could empower people to keep themselves healthy by encouraging them to make positive lifestyle changes to reduce risk, including through targeted healthcare nudges.
AIGHP for targeted intervention
Most proposed uses of polygenic scoring to target early interventions involve identifying those in the highest percentiles of risk for a disease. For instance, this is the application of polygenic scoring that Genomics PLC, which is a partner to Our Future Health, set out in its white paper on the potential value of polygenic scoring in UK healthcare.[135]
This use of polygenic scoring works in the following way:
- Combined risk scoring enables more accurate identification of those at high risk of a given disease, compared with conventional risk scoring approaches alone.
- Improvements in accuracy result in more people being identified who are genuinely at high risk of a disease. This is sometimes referred to as ‘reclassifying’ some people as high risk who were previously thought to be at low or average risk.
- Those reclassified into the high-risk category (who otherwise would have been missed) can be given an intervention that reduces their risk of developing the disease or prioritised for screening.
While this approach could deliver preventative benefit for those who are reclassified, its ability to significantly reduce overall healthcare demand is more uncertain.
Part of the reason for a potentially small impact on disease incidence is that for most common diseases, most cases across a population occur in those who are at low to moderate risk.[136] Conversely, those in the highest-risk groups are typically responsible for a small proportion of overall cases. This is known as the ‘prevention paradox’. It arises because those at low to moderate risk of a disease typically substantially outnumber those at high risk.[137] [138] [139] As this field develops, consideration should be given to the operation of the prevention paradox and whether AIGHP contributes to a better understanding of risk of disease, linked to appropriate interventions to prevent it, so that fewer cases occur in those currently considered to be at low to moderate risk.
AIGHP for targeted screening
Another potential use of AIGHP is to help target conventional screening programmes for common diseases, particularly cancer. In cases where screening is expensive or comes with other, non-monetary costs, those with high polygenic (or combined) risk scores could be prioritised for earlier or more frequent screening than those with low polygenic scores.[140]
This approach is argued to have two potential advantages over more wide-scale, less targeted screening: first, in the case of cancer, some screening programmes can result in overdiagnosis and overtreatment;[141] second, a more targeted approach to screening could potentially save money and resources by avoiding screening people at low risk of particular conditions.[142]
As with all screening programmes, developing a more targeted approach will involve complex trade-offs that will need careful evaluation to ensure that positive effects on those identified to be at higher risk are balanced against any negative impacts on those at lower risk.
AIGHP for lifestyle and behaviour change
Finally, AIGHP might help to reduce healthcare demand by bringing about positive behaviour and lifestyle change. Specifically, it has been suggested that AIGHP insight about relative disease risk could empower people to keep themselves healthy.[143] Armed with a clearer understanding of their genetic risk factors, people might be able to (and could be ‘nudged’ to) make more informed and healthier choices about how they live.
There is, however, patchy evidence to support the idea that better knowledge of disease risk leads to healthy behaviour change.[144] Moreover, current evidence regarding the ability of nudges to influence complex, long-term patterns of behaviour, such as those that determine health, is unclear.[145] Health-affecting behaviour and lifestyle choices have been shown to be influenced far more directly by environmental factors than by information about disease risk.[146] [147]
Any positive impacts of AIGHP insight on behaviour would also have to be carefully considered against unintended consequences of providing people with insight into their disease risk. In particular, it has been noted that unless they are communicated and explained clearly, providing people with their polygenic scores could give them a false sense of security in the case of being found to be at low or moderate polygenic risk.[148]
Once again, the prevention paradox comes into play: even if positive behavioural change improves outcomes for those at highest risk, the overall impact for common diseases across the whole population will likely still be low. The public health challenge of preventing common diseases remains.
The public’s views on fragility
While this was not a universal view, many participants were concerned that using AIGHP to shift the UK’s health systems towards a focus on prevention would place unrealistic expectations on individuals to keep themselves healthy. Many of these participants expressed scepticism regarding the ability of AIGHP to help most people stay healthy, and to meaningfully reduce demand for healthcare services.
Addressing gaps in law, regulation and governance
This chapter assesses the capacity of the UK’s current legal, regulatory and governance framework to address two of the risks identified in the previous chapter: surveillance and discrimination. It provides an overview of the current regulatory landscape relevant to the use of AIGHP systems in healthcare, identifies gaps and deficiencies in that landscape, and puts forward a series of proposals for strengthening existing protections.
The analysis and recommendations that we present in this chapter are informed by our research, including engagement with experts.
On protections against surveillance, we found that:
- existing data protection rules are unclear and err on the side of under-protecting the data required for AIGHP systems
- people have too little control over what is done with their genomic data once it is shared.
On protections against discrimination, we found that:
- the UK offers few protections against the kind of genomic discrimination that AIGHP systems would enable.
We put forward several recommendations for how these issues can be addressed, which are set out in order below and then further explored in the rest of the chapter.
Recommendations to address gaps in law, regulation and governance
Recommendation 1: Any future reforms of UK data protection law should stipulate that genomic data should always be considered personal data. This would constitute an important revision to the current, context-dependent definition of personal data in UK data protection law (which holds that genomic data is only personal data when identifiable). Such changes should be designed to avoid circumstances in which determining whether a genomic dataset is personal data requires knowledge of the capabilities of particular data processors.
Recommendation 2: Any future reforms of UK data protection law should clarify how to interpret the UK General Data Protection Regulation (GDPR) definition of healthcare data. This should be done in a way that complements the current approach of the UK GDPR, under which healthcare data is defined by its ability to reveal information about a person’s health.
Specifically, the law should be clarified to:
- provide additional detail on what counts and does not count as revealing information about a person’s health status – and especially on what counts as revealing information about a person’s mental health status
- specify that only data capable of revealing information about a person’s health status on its own, or in combination with a limited number of other data points, should be considered healthcare data.
Following any such reform, the Information Commissioner’s Office should consider producing guidance setting out common examples of kinds of data that do and do not count as healthcare data.
Recommendation 3: Any future reforms of UK data protection law should clarify that biometric data should be considered special category data in all circumstances, regardless of the primary purposes for collection.
Recommendation 4: The Department for Health and Social Care, the General Medical Council and other relevant organisations should work together to create a more granular model of consent under which subjects can specify in greater detail what they want to be done with data they share.
This model should be used for patients sharing their genomic data for research or clinical purposes and for research participants. It should provide a new set of standardised options that are structured to enable people to explicitly opt out of particular uses of data, including sharing data with particular entities. Future reforms to UK data protection law should state clearly that these explicit vetoes mean that the ‘compatibility test’ (which requires entities seeking to process special category data for a new purpose to demonstrate compatibility with the original purpose for which consent was given) is not passed.
Recommendation 5: The Department for Health and Social Care and the General Medical Council should conduct a deliberative public engagement exercise to inform the development of the new, more granular model of consent proposed in recommendation 4.
Recommendation 6: Any future reforms to UK data protection law should strengthen, rather than weaken, protections around the repurposing of genomic and phenotype data for research purposes. Specifically, for genomic and phenotype data, any future amendments should preserve:
- the ‘transparency requirement’ around repurposing of special category data (so that entities processing special category genomic and phenotype personal data for a new purpose are still obliged to inform the data subject, even where the data is being processed for the purposes of research)
- the ‘compatibility test’ around repurposing of special category data (so that entities seeking to process special category genomic and phenotype personal data for a new purpose still have to demonstrate compatibility with the original purpose for which consent was given, even where the data is being processed for the purposes of research).
Recommendation 7: The Government should develop an updated code of practice for the use of genetic and genomic data in the insurance industry. Building on the Association of British Insurers’ Code of Practice on Genetic Testing and Insurance, the new code should:
- prohibit the use of the results of predictive genetic and genomic tests for any kind of insurance, including life insurance, loss of earnings insurance and critical illness insurance
- explicitly define predictive genetic testing to include tests predicting both disease risk and drug responses, and to include testing looking at the risk of both genetic disease and common diseases
- define diagnostic genetic testing as applying to existing, symptomatic monogenic diseases, rather than common monogenic variants associated with disease risk; the latter should be explicitly considered predictive tests.
The Government should introduce primary legislation:
- requiring all insurers operating in the UK to comply with the updated code of practice
- enabling limited aspects of the code, such as monetary thresholds, to be amended by presenting the code before parliament, but making more substantive amendments to the code (including the kinds of genetic and genomics tests and insight an insurer may consider) impossible without the passage of new primary legislation.
Recommendation 8: The Government, the Equality and Human Rights Commission, relevant sector regulators and civil society should run a citizens’ assembly to explore the need for new primary legislation designed to address genomic discrimination, both in healthcare and in other domains, such as employment and education.
UK data protection law and medical and research consent practicesIn the UK, the main bodies of law, regulation and governance that most directly impact AIGHP and the risks associated with it are data protection regulations, and consent practices in medical research and clinical contexts. Data protection law: This imposes limitations on the circumstances under which identifiable genomic data can be collected and processed. The most relevant pieces of data protection legislation are the Data Protection Act 2018 (DPA), and the UK GDPR, which is the UK’s implementation of the EU GDPR. When interpreting UK data protection law, these two pieces of legislation must be read and considered in parallel. UK data protection law makes a distinction between three broad categories of data:
The DPA was set to be significantly amended by the Data Protection and Digital Information Bill, until the legislation was dropped in the run-up to the July 2024 UK general election. The proposed changes, and their implications for what could be done with data used for or produced by AIGHP systems, are discussed below in the context of potential future reforms to UK data protection law. Policy governing consent in medical contexts: This requires that genomic data be collected only with subject consent. These requirements are governed by the DHSC, the General Medical Council (in the case of consent in clinical practice), and the National Institute for Health and Care Research. Other entities (such as Genomics England) may issue further guidance on consent, as long as it is in line with the policy set out by the aforementioned entities. There is also a body of case law governing consent in medical contexts, with failure to obtain consent leaving medical practitioners and researchers open to the charge of clinical negligence. |
Further law and regulations relevant to AIGHPIn addition to data protection law and scientific and medical consent policies, several other laws and bodies of legislation apply to AIGHP. These include: The Human Tissue Act 2004: This Act makes it illegal to derive genomic data from human tissue without a subject’s consent. While this rules out some non-consensual uses of genomic data, it does not cover the analysis of genomic data that has already been collected. The Medicines and Medical Devices Act 2021: This Act takes a product safety approach to medical devices, dividing them into separate risk categories depending on the contexts in which and uses for which they are deployed. AI and software used in healthcare are subject to this framework, but it is as yet unclear how particular deployments of AI, including AIGHP systems, might fall within the risk classification. While the Medicines and Medical Devices Act will likely serve as an important regulatory constraint on how AIGHP systems are used, as a piece of product legislation it is unlikely to say much about preventing or constraining harms associated with privacy or lack of agency over genomic data on the part of data subjects. If the harms it identifies are construed very broadly, it may have something to say on issues of discrimination, but this is likely to be confined to questions of machine bias and differing rates of predictive accuracy for different groups, rather than addressing questions about the results of AIGHP systems being used to justify acts of discrimination. Human rights law: This sets out broad principles many of which may apply to the use of AIGHP in medical contexts. While UK human rights law is likely to apply to the use of genomic data and AIGHP systems, its specific implications for how the technology might be used are not straightforward and would be the subject of legal argument. Moreover, the two pieces of human rights legislation most directly related to AIGHP, the Human Rights Act 1998 (HRA) and the Equality Act 2010, have features that could limit their ability to constrain the use of AIGHP systems:
|
Problem: Inadequate protections against surveillance
One of the principal risks identified in the previous chapter was the possibility of AIGHP systems undermining people’s privacy and creating circumstances in which it is difficult for them to retain control over their personal (genomic and non-genomic) data.
Two of the most direct mechanisms by which the law can protect against these risks are (1) data protection regulation and (2) consent processes in medical governance. The following section highlights deficiencies in our existing protections and sets out recommendations for addressing them.
Driver: Data protection rules are unclear and err on the side of under-protection
One clear finding from our expert interviews was that the UK’s current data protection laws provide policymakers and regulators with most of the high-level tools needed to address potential privacy and surveillance harms posed by AIGHP systems. However, there are several open questions about how UK GDPR and the 2018 Data Protection Act might apply to AIGHP in specific instances.
We argue that reforms to UK data protection law are now needed to clarify longstanding questions about how and in what circumstances UK data protection law applies to genomic and phenotype data. As AIGHP raises both the stakes and the frequency of genomic and phenotype data collection, the need for unambiguous guidance on its collection and processing is likely to become more pressing – and the absence of such guidance more problematic.
Addressing these ambiguities matters. Data protection law is one of the most direct and powerful mechanisms available to provide individuals with control over if and how their genomic and phenotype data is used, and with a degree of protection against decisions made by systems using this data. A lack of clarity regarding how such rules apply to data required for AIGHP systems dramatically undermines their efficacy. Specifically, ambiguity means that:
- the public have no real way of knowing what can and cannot be done with their genomic and phenotype data, or who this data might be used and held by
- those collecting and processing data for AIGHP systems can do so with greater impunity than if the law were clear
- it is difficult for researchers to know how best to treat genomic data, and this can in some cases represent an impediment to research.
It is unclear when genomic data is covered by UK data protection law
UK data protection law explicitly states that all genomic personal data counts as special category data.[149] However, in many cases it can be difficult to determine when genomic data should be counted as personal data. As a consequence, it is not always clear whether a given genomic dataset should enjoy the highest degree of protection under UK data protection law (special category data status), or whether it fails to fall under the remit of data protection law at all. The problematic nature of this ambiguity in the context of genomic data is evidenced by research conducted in 2020 by the PHG Foundation, which found that professionals were ‘experiencing challenges reaching consensus about when genomic and associated health data are “personal data”’.[150]
The basis of the ambiguity
UK data protection law defines personal data as ‘any information relating to an identified or identifiable living individual’. For it to count as personal data, it has to be possible to use that data to directly or indirectly identify a person.[151] The problem is that it is unclear what it takes for a person to be identifiable from a given piece of data. There are at least two competing ways to interpret this test, both of which present difficulties.
One way is to ask whether it would be possible to identify a person from the data in question under any circumstances. A difficulty with this is that practically any data could conceivably enable this when combined with enough other data and subjected to sufficiently sophisticated analysis. Interpreting personal data in this way could therefore end up counting almost all forms of data about living people as personal data.
An alternative is to ask whether it would be practically possible to identify a person from a given piece of data, given current technological and data constraints. While this has the advantage of not automatically counting all data about living people as personal data, it requires a clear and shared understanding of what is currently technologically possible, and what other data might theoretically be available to enable an identification. It also means that what counts as personal data is liable to change as technological capabilities and data availability evolve.
These challenges are particularly acute in the case of genomics, where there are controversies about the ability of current techniques to identify individuals, and where technological capabilities and the availability of complimentary datasets are developing rapidly and unpredictably.
Previously proposed changes would not have solved the problem
The Conservative Government’s Data Protection and Digital Information Bill (DPDI), which was one of several pieces of legislation that failed to pass parliament when a general election was called in May 2024, included changes intended to clarify the correct interpretation of identifiable data under UK data protection law. In the King’s Speech in July 2024, the new Labour Government announced plans for a Digital Information and Smart Data Bill that may bring back several aspects of the former Bill.[152] It is therefore important to set out why the changes proposed in the original DPDI Bill would have failed to address ambiguities around when genomic data counts as personal data.
The Bill specified that a data controller or processor should consider data identifiable (and therefore personal data) if they, or others they reasonably expect to have access to that data, are able to identify a living person from that data ‘by reasonable means’.[153]
In one sense, this would have addressed part of the ambiguity in UK data protection law. The question of whether the test for personal data is ‘Is it possible to identify a person from this data?’ or ‘Is it possible for anyone likely to have access to this data to easily identify a person from it?’ would have been definitely resolved in favour of the latter.
However, although this would have made it clearer what the question is, it would not have made that question any easier to answer. Indeed, in order to determine whether data is personal, it is necessary to understand who is likely to get hold of it and the technological, financial and other resources they may have at their disposal to identify an individual from it.[154]
Answering these questions requires far more assumptions and specific knowledge – and is therefore far harder to do – than answering the question of what could be done with a dataset given its intrinsic properties. Moreover, these questions are especially difficult to answer in the case of genomic data, where datasets can be shared and reshared widely, where very small amounts of genomic data can be used to identify a natural person, and where the technology and techniques used to identify and de-identify individuals are advancing rapidly.[155]
Proposed solution: Err on the side of all genomic data counting as personal data
In addressing the difficulties in determining whether genomic data should count as personal data, it is necessary to choose between two imperfect solutions. One is to take a contextual approach, attempting to reflect the nuances of current genomic science and the position and abilities of those holding the data, setting out exactly the criteria for when genomic data should be considered identifiable and when it should be considered unidentifiable. The other is to state, for the sake of simplicity and universality of protection, that genomic data should always be considered special category data.
Our view is that the latter is preferable. Erring on the side of inclusion would provide data subjects with far more robust protection as technology develops – and would avoid the risk of genomic data that has not been categorised as needing special category treatment becoming identifiable after the fact. Moreover, having to work out when genomic data does and does not count as special category data is likely a bigger impediment than a clear requirement to always treat it as special category.
Recommendation 1: Any future reforms of UK data protection law should stipulate that genomic data should always be considered personal data. This would constitute an important revision to the current, context-dependent definition of personal data in UK data protection law (which holds that genomic data is only personal data when identifiable). Such changes should be designed to avoid circumstances in which determining whether a genomic dataset is personal data requires knowledge of the capabilities of particular data processors.
Problem: it is unclear when phenotype data counts as special category data
If individuals are to have any control over the kinds of AIGHP research for which their data is used, or the AIGHP systems to which they may be subjected, they need control over their phenotype data as well as their genomic data.[156]
In practice, providing individuals with an appropriate degree of control over their phenotype data requires treating phenotype data used for AIGHP as special category data.
Under current UK data protection, however, identifiable phenotype data is not automatically considered special category. Moreover, there is considerable ambiguity about when identifiable phenotype data does count as special category data.
Phenotype personal data counts as special category data when it is considered (1) health data or (2) biometric data used to identify a natural person. Future reforms to UK data protection must address the considerable ambiguity about when either of these categories apply.
Driver: It is unclear when phenotype data counts as health data
Under GDPR, ‘“data concerning health” means personal data related to the physical or mental health of a natural person, including the provision of healthcare services, which reveal information about his or her health status’.[157]
Interpreting what kinds of data actually fall under this definition is fraught with difficulty. A lot of weight is placed on the question of what counts as revealing information about a person’s health status. There are open questions about how this criterion should be interpreted: does data have to be capable of revealing information about a person’s health status on its own, or in combination with other data? How immediate does the connection between the data and the health status need to be, and with what degree of certainty do inferences need to be made?
These questions bear directly on whether phenotype data counts as healthcare data: while phenotype data includes things obviously related to and collected for the purposes of health and healthcare, such as medical records or the results of diagnostic tests, it also includes datapoints whose status is more ambiguous, such as data collected from wearables and elements of a person’s ‘digital phenotype’.[158] It could also include things that do not appear traditionally related to health, such as a person’s level of education, social habits or employment status, but which correlate to health current and future health status.
Debates on what counts as health data
The question of where (if at all) the line between health data and non-health data should be drawn, and how difficult cases should be treated by European law, has a long history that predates the GDPR.
For instance, in response to a question about how data from wearables and lifestyle apps should be interpreted under the EU Data Protection Directive (the predecessor to the GDPR), the EU Article 29 working group argued that in European law, health data should be interpreted broadly. The working group identified three main scenarios in which data collected by lifestyle and wellbeing apps and devices constitutes health data, two of which as are follows: ‘The raw sensor data processed by the app or device can be used, independently or in combination with other data, to draw conclusions about an individual’s actual health status or health risks’ and ‘The data allows for conclusions to be drawn about an individual’s health status or health risks (irrespective of whether these conclusions are accurate or inaccurate, legitimate or illegitimate or otherwise adequate or inadequate)’.[159]
While this might appear to provide clarity, the working group’s interpretation of health data is very broad, rendering practically any identifiable personal data potentially health data.[160] Rather than settling the question of what counts as health data, the intention of the working group appears to have been to provide members states’ data protection authorities (the ICO in the case of the UK) with the maximum amount of latitude in protecting data subjects’ rights under European law. Critically, the fact that the ICO could choose to interpret the Data Protection Directive (and by extension the GDPR) this broadly does not mean that they must, or indeed that they do.
The ICO does not have any guidance on how the GDPR definition of health data should be understood, and current legislation does provide an indication of how these ambiguities should be resolved. As such, what counts as health data in UK data protection law, and how phenotype data edge cases should be dealt with, remains unclear.
Proposed solution: Future reforms to UK data protection law should make clearer what does and does not count as health data
UK data protection law needs to offer additional clarity on when phenotype data should be considered healthcare data.
A challenge here is to develop regulation that reflects the increasingly large amount of data types that could be considered relevant to health and healthcare, but that avoids counting all personal phenotype data as healthcare data.
One option is to focus on the purposes for which data is processed, with only phenotype data collected or processed for the purposes of healthcare provision or research being counted as healthcare data. While this would have the advantage of excluding the huge amount of personal data with merely potential relevance to health, it could present enforcement challenges. In particular, some experts with whom we engaged suggested that a ‘purpose-based’ definition could potentially allow a data controller to collect and process phenotype data for non-health purposes but then use that data to make inferences about people’s health once collected.
A potentially superior approach would be to distinguish between phenotype data that could reveal information about a person’s physical health on its own (or in combination with a small number of other data points) and phenotype data that could reveal information about a person’s physical health only in combination with large numbers of other data points. Legislation could stipulate that, all else remaining equal, only the former should be considered healthcare data. While the exact threshold between these two kinds of phenotype data may have to be set somewhat arbitrarily, this approach would help provide a way of avoiding a situation in which practically all data could be argued to be healthcare data.
Recommendation 2: Any future reforms of UK data protection law should clarify how to interpret the UK General Data Protection Regulation (GDPR) definition of healthcare data. This should be done in a way that complements the current approach of the UK GDPR, under which healthcare data is defined by its ability to reveal information about a person’s health.
Specifically, the law should be clarified to:
- provide additional detail on what counts and does not count as revealing information about a person’s health status – and especially on what counts as revealing information about a person’s mental health status
- specify that only data capable of revealing information about a person’s health status on its own, or in combination with a limited number of other data points, should be considered healthcare data.
Following any such reform, the Information Commissioner’s Office should consider producing guidance setting out common examples of kinds of data that do and do not count as healthcare data.
Driver: It is unclear when phenotype data counts as biometric data
The UK GDPR defines biometric data as ‘personal data resulting from specific technical processing relating to the physical, physiological or behavioural characteristics of a natural person, which allow or confirm the unique identification of that natural person, such as facial images or dactyloscopic data’.[161]
Under this definition, many kinds of phenotype data likely to be collected and processed for the purposes of AIGHP count as biometric personal data. However, the UK GDPR states that biometric data counts as special category only when it is collected or processed with the intention of identifying a natural individual.[162]
A problem with this requirement is that it fails to afford special category protection to biometric data used to produce AIGHP insight about people. Biometric data collected about a subject for the purposes of generating a genomic prediction about that person (such as a behavioural trait or intelligence), rather than for the purposes of identifying that person, would not necessarily have to be treated as special category data.
Proposed solution: UK data protection law should be reformed to clarify what biometric data counts as special category data
In contrast to healthcare data, the question of what counts and does not count as biometric data is reasonably clear-cut under UK data protection law. It would be possible simply to specify that all biometric data should count as special category, rather than only biometric data collected for the purposes of identification. This would lead to more forms of biometric data counting as special category and would avoid the problem of biometric data collected for the purposes of categorisation (and AIGHP) not being afforded special category status. It would therefore provide data subjects with much more control over a key dataset feeding into AIGHP systems.
Recommendation 3: Any future reforms of UK data protection law should clarify that biometric data should be considered special category data in all circumstances, regardless of the primary purposes for collection.
Driver: People have too little control over what is done with genomic data that they share
Another, broad problem is the low level of control that people have over how genomic data shared for research is used and reused.
It is common for data collected for the purposes of scientific or medical research to be reused for further research projects. Likewise, genomic data collected in the course of clinical practice (for example, for diagnostics or to inform treatment decisions) and by direct-to consumer genetic testing[163] can be shared for the purposes of scientific medical research.
In these cases, the sharing and repurposing of genomic data is governed by consent and data protection rules. Medical and research ethics requires that people participating in research or undergoing treatment consent to their data being shared with third parties and to that data being used for new purposes. Similarly, under UK data protection law, when an entity processes genomic data for a new purpose it must either seek new consent from the subject or demonstrate that the new purpose is compatible with the original one for which consent was obtained.
While these rules ensure that a person’s genomic data cannot legally be collected and shared without explicit consent, they provide people with little de facto control over what is done with their genomic data once it is shared.
Consent rules do not allow data subjects to specify their preferences in much detail
One reason for this is that it is rare for consent processes to enable subjects to specify their preferences regarding processing in any level of detail. While there is considerable variation between consent processes in scientific and medical research, those in clinical settings, and those for commercial entities (such as direct-to-consumer genetic testing companies), research subjects typically get a choice about whether to share their data for a specific purpose (specific consent) or for a more broadly defined purpose (wide consent). Though there are moves to make consent in these settings more granular and less of a one-off decision, such practices are not universally required and are not yet widespread.
| Granular consent
In the context of consent processes for data sharing and research, ‘granularity’ refers to the degree of precision with which subjects are able to specify how they want their data to be used. A granular consent process may, for instance, enable a research or data subject to specify the exact uses or kinds of purposes for which their data may and may not be used and the specific entities or kinds of entities who may use and may not use the data. By contrast, less granular or non-granular consent processes require individuals to accept or reject the terms of how their data may be used as a bundle, with far more limited ability to pick and choose if, how and where their data is used.
Dynamic consent This refers to a flexible approach to managing consent for data use, generally facilitated by digital platforms. Dynamic consent allows those sharing their data to modify their consent preferences over time as their circumstances or preferences change. |
Data protection rules do not enable people’s consent preferences to constrain what is done with genomic data
This problem is compounded by the fact that the purposes for which a subject provides consent exert a tenuous, difficult-to-predict influence on the new forms of processing that a processor can legally conduct. A processor does not generally need to seek fresh consent to process a subject’s data in a new way. Instead, they can argue that the new purpose is not incompatible with the purpose for which consent was originally given, or they can cite one of the alternative legal bases for processing special category data outlined in the UK GDPR.[164]
Assessing the compatibility of a new purpose for processing with the original purpose is not straightforward, and the expectation appears to be that assessments should be made on a case-by-case basis, with the concept of compatibility to be interpreted broadly. Opinion 03/2013 on purpose limitation in European data protection law (which is the original source of the UK GDPR) notes that the original law was likely written so as to provide a degree of flexibility and openness with regard to how data can be repurposed in the future.[165]
Critically, the wording of the test as ‘not incompatible’ could suggest that the absence of a data subject’s active consent for a particular purpose does not imply that that purpose is incompatible with the original purpose. For a new purpose to be incompatible for this reason, the data subject would presumably have to actively reject it in their original consent statement.
The upshot of this broad interpretation of compatibility is that it is hard for a data subject to know what uses may be considered compatible with those to which they have consented.
Proposed changes would make this problem worse
The previous Conservative Government’s Data Protection and Digital Information Bill also included changes that would have made it easier for researchers and scientists to repurpose personal data. Reforms of the kind proposed would further weaken the ability of consent processes and data protection law to constrain the repurposing of genomic data. It is possible that Labour’s forthcoming Digital Information and Smart Data Bill may try to incorporate similar provisions.
The previous Bill stated that in cases of scientific research, compatibility with the original purpose to which consent was given should be assumed.[166] The Ada Lovelace Institute commissioned the legal consultancy AI Law to produce a legal analysis of the Bill, which found that it would enable personal data to be used in ways that the data subject might not anticipate.
The Bill also stated that in cases of scientific research, a data processor would no longer have any obligation to inform the data subject when their data was being processed for new purposes.[167]
In addition to depriving data subjects of information about how their data was being used, this change would have made it far more difficult for them to withdraw consent for data processing. Currently, a data subject can withdraw consent if it transpires that their data is being used for purposes which they do not consent to. If the proposed changes had come into force, there would have been far less legal obligation to provide data subjects with the information they would need to inform such decisions.
These provisions would have removed the data subject’s right to obtain transparency about when their data was being repurposed by the controller who originally collected it. This would have weakened a data subject’s rights in relation to their data.[168]
There are several questions about how these changes would have operated in practice: it is unclear whether this exemption from the compatibility test in cases of scientific research would still hold if consent processes explicitly ruled out particular uses of data. There is also a question of whether the two changes referred to above, by linking exemptions to ‘scientific or historical research’, would have made this category more consequential than it has been previously – with questions of what counts as scientific or historical research becoming more important and contested than otherwise.
Proposed solution: Consent should be made more granular, and data protection law should be amended to ensure that consent is better able to constrain what is done with genomic data
Data subjects’ low degree of control over what is done with their genomic data once it is shared is a substantial problem. The inability of subjects to specify the purposes for which they are happy to share their genomic data, and their inability to have their preferences reflected, turns the decision of whether to share genomic data into an unhelpfully all-or-nothing choice: either subjects share their data for research purposes without knowing how else it could be used, or they decide to withhold it completely. This significantly undermines people’s ability to use existing legal and governance mechanisms to maintain their genomic privacy and protect themselves from harms, such as discrimination, that might arise from their data falling into the wrong hands.
It is likely that the inability of data subjects to control what is and is not done with their genomic data will, in the long run, impact the number and kinds of people willing to participate in genomic research. There is compelling evidence, from public engagement and from data scandals over the past decade, that a significant proportion of the UK public care about their privacy when sharing their genomic data and have preferences about the kinds of research (and research entities) to which they do and do not want to contribute.[169] Transparency by those using data and engagement with subjects will be key to sustaining public acceptability even in the context of appropriate permissions and consents.
Given that researchers will have to be open about the options available to research and data subjects, it is likely that the inability of subjects to set the terms of data sharing will be a significant impediment to increasing the participation of under-represented groups in genomics research.
Recommendation 4: The Department for Health and Social Care, the General Medical Council and other relevant organisations should work together to create a more granular model of consent under which subjects can specify in greater detail what they want to be done with data they share.
This model should be used for patients sharing their genomic data for research or clinical purposes and for research participants. It should provide a new set of standardised options that are structured to enable people to explicitly opt out of particular uses of data, including sharing data with particular entities. Future reforms to UK data protection law should state clearly that these explicit vetoes mean that the ‘compatibility test’ (which requires entities seeking to process special category data for a new purpose to demonstrate compatibility with the original purpose for which consent was given) is not passed.
Recommendation 5: The Department for Health and Social Care and the General Medical Council should conduct a deliberative public engagement exercise to inform the development of the new, more granular model of consent proposed in recommendation 4.
Recommendation 6: Any future reforms to UK data protection law should strengthen, rather than weaken, protections around the repurposing of genomic and phenotype data for research purposes. Specifically, for genomic and phenotype data, any future amendments should preserve:
- the ‘transparency requirement’ around repurposing of special category data (so that entities processing special category genomic and phenotype personal data for a new purpose are still obliged to inform the data subject, even where the data is being processed for the purposes of research)
- the ‘compatibility test’ around repurposing of special category data (so that entities seeking to process special category genomic and phenotype personal data for a new purpose still have to demonstrate compatibility with the original purpose for which consent was given, even where the data is being processed for the purposes of research).
Problem: Lack of protections against discrimination
A recurring concern from our expert interviews and public engagement is that AIGHP could enable more forms of discrimination against certain individuals or groups, particularly those who have been traditionally marginalised in healthcare. The most cited risk for discrimination in our public engagement exercise was the use of insights from AIGHP to determine what kinds of health insurance an individual is offered.
As set out in the previous chapter, the ability of insurers to discriminate on the basis of the results of genomic disease risk prediction tools could have several adverse consequences. Public anxiety about these possibilities also presents a significant obstacle to widening and diversifying participation in genomic research and medicine, and there are strong reasons to believe that this problem could get worse.
The problem is particularly acute for the UK, a country with especially ambitious plans for the use of genomics in medicine and research which would require a dramatic expansion in public participation in genomic testing.[170] More significantly, failure to take robust action on genomic discrimination could lead to outcomes in which those most in need of healthcare struggle to access it affordably.
Driver: The UK has few real protections against the kind of genomic discrimination that AIGHP systems would enable
The UK has poor protections against genomic discrimination and the use of AIGHP insight by health (and other) insurers. As a consequence, it has relatively few legal and regulatory protections to reassure those worried about the consequences of sharing their genomic data.
The UK’s current protections against genomic discrimination
Arguably the most significant piece of UK legislation pertaining to discrimination is the 2010 Equality Act. The Act provides protection against discrimination on the basis of nine protected characteristics: age, disability, gender reassignment, marriage and civil partnership, pregnancy and maternity, race, religion or belief, sex, and sexual orientation.[171]
The Equality Act considers an action or activity discriminatory only if it discriminates on the basis of one of these protected characteristics. This makes it poorly suited to addressing discrimination by genomic analysis (and therefore by AIGHP). By definition, genomic discrimination is discrimination on the basis of a person’s genotype. Genomic discrimination might, for instance, involve offering a person a more expensive insurance premium because they have a specific combination of genetic variants that suggest they have a heightened risk of developing a particular kind of cancer.
There may be some genomic variations that are disproportionately associated with possession of a particular protected characteristic. In these cases, it may be possible to argue that acts of genomic discrimination on the basis of those traits are contrary to the Equality Act, because such discrimination amounts in practice to discrimination against people with a protected characteristic.
However, this provides patchy, contingent protection against genomic discrimination. The majority of instances of genomic discrimination will be on the basis of specific sets of genomic variations that, while relevant to some phenotypic trait, do not correspond to any pre-existing categories or groups of people. For instance, a person’s increased risk of a particular kind of cancer might be the consequence of a particular set of genomic variants, possession of which does not correlate to possession of any other protected characteristic (such as race, sex or disability).
One potential solution would be to add genotype to the list of protected characteristics. A potential – though presumably not insurmountable – challenge with this approach would be how to clearly distinguish between instances of discrimination on the basis of genotype and on the basis of phenotype.[172]
Alongside the Equality Act, the 1998 Human Rights Act contains provisions that are indirectly relevant to discrimination, which could apply to cases of genomic discrimination. Most notably, Article 14 of the Act states that the enjoyment of the rights and freedoms set out in the Act (which are those of the European Convention on Human Rights (ECHR)) ‘shall be secured without discrimination on any ground such as sex, race, colour, language, religion, political or other opinion, national or social origin, association with a national minority, property, birth or other status’.[173] However, other than the stipulation that the rights set out in the law should be secured and applied in a non-discriminatory manner, the HRA includes no specific provisions on discrimination, let alone any specific provisions on discrimination on the basis of genotype.
The consequence of this is that the Human Rights Act cannot be presumed to – and does not automatically – offer protections against the kinds of genomic discrimination identified in the report. For a given instance of genomic discrimination to be contrary to the Human Rights Act, it would have to be actively argued and established that the instance impeded the enjoyment of one of the rights or freedoms of the ECHR. Moreover, while the inclusion of the words ‘discrimination on any ground’ and ‘or other status’ means that, unlike the Equality Act, the Human Rights Act can cover cases of discrimination on the basis of characteristics other than the ones it explicitly lists, this does not guarantee that the law would be interpreted as covering discrimination on the basis of genotype.
In addition to being general legislation whose relevance to specific instances of genomic discrimination would have to be established by legal argument, the Human Rights Act is limited to public bodies and those carrying out public tasks and duties, and does not apply to the private sector or to individuals.
The inability of the UK’s equalities and human rights laws to directly address genomic discrimination is compounded by a lack of legislation and explicit commitments on the issue. Notably, the UK has not signed up to the Oviedo Convention (the European Convention on Bioethics), an international instrument prohibiting the misuse of innovations in biomedicine and ensuring the protection of human dignity. The Oviedo Convention proscribes any form of discrimination against a person on the grounds of their genetic heritage.
The UK also lacks any domestic legislation directly addressing genomic discrimination, or the use of genomic testing and prediction, in the context of insurance. Many states, including the USA, Canada, France, Germany, the Netherlands and Austria, have passed legislation explicitly prohibiting discriminatory uses of genetic and genomic analysis and of genetic testing in the insurance industry.
| Legislation addressing genetic discrimination and the use of genetic testing in insurance[174]
United States: The Genomic Information Non-Discrimination Act (GINA) bars the use of genetic information in health insurance and employment decisions. Canada: The Genetic Non-Discrimination Act 2017 makes it illegal to require an individual to undergo or disclose the results of a genetic test as a condition of entering into a contract, or the provision of goods or services. France: Law 94-653 on the respect of the human body and 2004-800 on bioethics both prohibit the use of genetic tests by insurers. Germany: The Diagnostics Act 2009 restricts the use of genetic testing to medical purposes and explicitly forbids its use in insurance contexts, expect for very high coverage amounts. The Netherlands: The Medical Examination Act limits the use of genetic information by insurers. Austria: The Gene Technology Act prohibits the use of the results of genetic tests by insurers and employers. |
By contrast, the UK has so far opted for a voluntary code, referred to as the ABI Code on Genetic Testing and Insurance. Under this arrangement, the UK insurance industry voluntarily commits to refrain from using genomic testing in all but a very limited set of circumstances.[175] While the Code is often cited as the UK’s equivalent to the legal protections that exist in other countries, it has two substantial weaknesses compared with these other regimes.
- The ABI Code does not legally prevent an insurer from engaging in genomic discrimination. Although signing up to and compliance with the terms of the Code is a condition of ABI membership, there is nothing compelling a UK insurer to be a member of the ABI. The existence of UK insurers who have not signed up to the Code is tacitly acknowledged on the Government’s website: ‘Most insurance companies who are not members of the ABI have also signed up to the code’.[176]
- The ABI Code is open ended, meaning that it could be revised in the future to enable insurers to take into account the results of genomic tests in ways that are currently ruled out. The open-ended nature of the code undermines its utility as a means of protecting people against genomic discrimination and of providing reassurance to those concerned about participation.
Proposed solution: The ABI Code of Practice on Genetic Testing and Insurance should be transposed into the UK statute book
The Code is often billed as a flexible, consensus-based approach to governance, which balances protection and reassurance for the public with the needs of the UK insurance industry in the face of fast-evolving medical science. On the latter point, the Government’s webpage on the Code states that ‘The code on Genetic Testing and Insurance aims to provide reassurance to the public about how and whether genetic testing could affect their access to certain types of insurance in the UK’.[177]
In reality, however, the non-binding, open-ended nature of the Code means that it fails to provide robust, future-proof protection against the use of genetic testing by insurers. It also fails to provide the public with genuine reassurance about how their genetic information might be used. Protections that can be ignored, and which may be repealed in the future, are no protections at all.
While the voluntary, open-ended nature of the Code could be argued to have been a proportionate response to the risk of genomic discrimination in the past – when AIGHP and other systems were still in their infancy, and health insurance played a very small role in the UK’s healthcare mix – the time is right to revisit this arrangement.
In the face of emerging technologies such as AIGHP and a rapidly expanding private healthcare sector, the status of the code is the single biggest deficiency in the UK’s regulatory protections against genomic discrimination. Given the material risk presented by the use of genomic information in insurance, public fears regarding genetic discrimination, and the impediment these present to widening participation in genomic medicine and research, there is a strong case for transposing the provisions of the code into the UK statue book. In addition to providing much-improved protection against the emergence of harmful practices, and genuine reassurance for the public, upgrading the code would provide certainty for investors and innovators that the UK’s governance approach is not liable to suddenly change.
We address common arguments for keeping the ABI code voluntary and open ended in Annex I.
Recommendation 7: The Government should develop an updated code of practice for the use of genetic and genomic data in the insurance industry. Building on the Association of British Insurers’ Code of Practice on Genetic Testing and Insurance, the new code should:
- prohibit the use of the results of predictive genetic and genomic tests for any kind of insurance, including life insurance, loss of earnings insurance and critical illness insurance
- explicitly define predictive genetic testing to include tests predicting both disease risk and drug response, and to include testing looking not just at genetic disease risk but also risk of common diseases
- define diagnostic testing as applying to existing, symptomatic monogenic diseases rather than common monogenic variants associated with disease risk. The latter should be explicitly considered predictive tests.
The Government should introduce primary legislation:
- requiring all insurers operating in the UK to comply with the updated code of practice
- enabling limited aspects of the code, such as monetary thresholds, to be amended by presenting the code before parliament, but making more substantive amendments to the code (including the kinds of genetic and genomics tests and insight an insurer may consider) impossible without the passage of new primary legislation.
Recommendation 8: The Government, the Equality and Human Rights Commission, sector regulators and civil society should run a citizens’ assembly to explore the need for new primary legislation designed to address genomic discrimination, both in healthcare and in other domains, such as employment and education.
Health policy and competing visions for AIGHP
The previous chapter identified gaps in current law, regulation and governance that need to be addressed to improve protection against the risks associated with the use of AIGHP in UK healthcare.
While we set out several concrete steps that could improve existing regulatory and legal protections, a clear message from the previous chapter is that the presence of robust regulatory and legal protections is a necessary but not sufficient condition for adequately addressing the risks posed by AIGHP. In many cases, the nature of the risks identified will also be determined by the choices made about how to deploy AIGHP in the UK’s healthcare system.
This chapter considers different strategic approaches that the NHS and UK policymakers could take to AIGHP. On the basis of the evidence we have gathered on the risks of AIGHP use, and the pressures and constraints that the NHS is operating under – and for reasons that we set out below – we make the following recommendations.
Recommendation 9: The Government, civil service and NHS should work to enable responsible, situational and high-impact deployments of AIGHP within the UK healthcare system. Such deployment should only be permitted where:
- adequate regulatory safeguards against surveillance and discrimination are introduced; gaps in data protection and anti-discrimination law covered in this report and in the previous recommendations must be addressed in advance of any deployment of AIGHP systems in the NHS
- the accuracy and reliability of AIGHP systems for different demographic groups reliably reaches a certain threshold; in its work on software and AI as a medical device, the Medicines and Healthcare Products Regulatory Agency should develop minimum standards of accuracy and efficacy for AIGHP systems and require any systems deployed in healthcare settings to meet them
- the NHS is demonstrably capable of and has committed to providing adequate and timely support for those who would be subject to AIGHP insight: any plans for deploying AIGHP in the NHS need to take account of the availability of genomic counselling for those subject to AIGHP insight; where the availability of genomic counselling is too low to provide it to everyone using AIGHP, and where there is no credible plan to expand access, AIGHP should not be deployed.
Where these conditions can be met, the Government and the NHS should work to enable the deployment of high-quality, carefully monitored AIGHP systems. To maximise impact, and to avoid cases where money and resources could deliver greater benefit elsewhere, AIGHP deployments should be restricted to cases in which there is a clear, clinically determined need for the extra insight provided by AIGHP, and where this benefit would outweigh any social and ethical risks, including discrimination and threats to privacy.
Recommendation 10: Given the risks and uncertainty about the accuracy and ability to reduce healthcare demand of AIGHP, the Department for Health and Social Care and the NHS should rule out the widespread deployment of AIGHP unless and until these uncertainties are resolved.
The Government, civil service and NHS should put in place safeguards to ensure that investments in uses of AIGHP are limited to those that are well evidenced, strategic and cost effective.
In funding, investment and resource allocation decision making and strategy, the NHS and Government should prioritise improving environmental determinants of healthcare outcomes over providing the whole population with insight into genomic variations in disease risk.
Any investments in AIGHP at scale for prevention should only be made where:
- this can be done in addition to, rather than in place of, addressing more fundamental problems with the health service
- there is clear evidence that providing of AIGHP to a large section of the population would result in significant and lasting reductions in demand for healthcare that could not be achieved more cost effectively through other interventions and investments
- concerns about privacy and individual control of genomic and healthcare data can be adequately addressed, and AIGHP can be rolled out so participation is optional rather than a de facto requirement of receiving adequate healthcare.
We make these final recommendations because as AIGHP technologies develop, the NHS will have to make strategic choices about how it wants to use this technology. As long as there is a chance of AIGHP being viable, the NHS will need a plan for how to deploy it. The NHS and the Government have invested money in AIGHP, and there is likely to be pressure to use it.
As with all new technologies, it might be tempting for policymakers to view AIGHP as a technological solution to deeper, more longstanding problems with the NHS.
A specific picture of how AIGHP might be used to transform UK healthcare is already taking form. Rapid advances in genomics and AI over the past five years have spurred an emerging narrative about how AIGHP could make the NHS far more preventative, automated and streamlined.
Pursuit of this vision of AIGHP is partly a gamble. It would require a very large, speculative investment in data collection, storage, and processing infrastructure and capacity. In some forms, it would amount to a fundamental redesign of how the NHS operates. One finding from the literature on the economic theory of technology is that new technologies start to deliver benefit only when new processes are developed to make use of them.[178] [179]
We believe that this vision for AIGHP in the NHS is risky, given the challenges with AIGHP that we have identified and the practical constraints to integrating new technological systems into the NHS.
We also believe that there are other ways for the NHS to use AIGHP that represent a better balance of the benefits, risks and costs of the technology, and which better manage the underlying uncertainties regarding its efficacy – as well as other ways of addressing some of the structural problems facing the NHS.
This chapter recaps the risks associated with AIGHP and addresses some of the practical challenges to implementing it in the NHS. We then compare the emerging, mass-prevention-oriented vision of AIGHP with an alternative and consider how the two perform with regard to the risks they pose, the benefits they bring and what they would cost.
Challenges to incorporating AIGHP into the NHS
Participants in our expert interviews and workshops highlighted several practical difficulties with incorporating AIGHP into the NHS.
Infrastructure requirements
Building and maintaining the necessary infrastructure for AIGHP would be a big task and is likely to have to be led by Government. Incorporating AIGHP capacity into the NHS at a large scale would require significant changes to data collection, storage and processing infrastructure. In our expert interviews, several participants suggested that the expansion of this capacity could only be carried out successfully through a large, Government-run project.
- The complexity and scale of the infrastructure and services required would make it difficult for the NHS to procure these services from a third party.
- It would be challenging to ensure a dynamic market for the provision of AIGHP services and to avoid vendor lock-in. The implications of procurement processes failing to deliver would be heightened because AIGHP commissioning would probably have to be centralised, as it would be too complicated to carry out at locally. For instance, it would be difficult, if not impossible, for an Integrated Care Partnership[180] to commission an AIGHP tool.
Workforce requirements
Our expert interviews also highlighted workforce concerns. AIGHP systems will be difficult to incorporate into a time-pressed NHS workforce. Clinicians either would need training in how AIGHP systems work and how to interpret results or would have to follow guidance on how to incorporate AIGHP insight into clinical decision making. One concern raised by our expert interviews was that the NHS workforce has little time for additional training of this kind. As a result, there is a risk that AIGHP systems may have to be deployed in a way that gives clinicians little ability to interrogate or understand the recommendations they are receiving.
Monitoring requirements
Finally, assessing the ongoing accuracy and efficacy of AIGHP systems will be difficult. The experts we engaged with highlighted difficulties in ensuring that tools based on prediction are sufficiently accurate.
To ensure that AIGHP insight is a legitimate guide to clinical and personal decision making, its performance will have to be carefully monitored, which will require the collection and processing of large amounts of clinical and healthcare data over long periods of time.
This task of monitoring the accuracy and efficacy of AIGHP systems will be made more difficult because predictions regarding future health are dynamic. AIGHP predictions are likely to be made using genomic data and health and lifestyle information, of which only the former will remain fixed over time. As such, the accuracy and utility of predictions may degrade over time. Moreover, the act of making a prediction can invalidate it. For instance, telling a person that they are at high risk of developing cardiovascular disease might prompt them to change their behaviour, thereby influencing their risk of developing the disease.[181] While this would be a beneficial outcome, it will require the NHS to expend additional resources to monitor AIGHP performance and ensure its thresholds for accuracy are continuously met.
Two competing visions of AIGHP use in the NHS
As illustrated by our scenario-mapping exercise, there are many ways that AIGHP could be incorporated into healthcare, each of which has a different mix of risks and benefits. Below, we set out two alternative visions for AIGHP in the NHS and assess them against what we know about the risks associated with AIGHP and the challenges of integrating new technologies into the health service.
A vision of AIGHP for mass prevention and healthcare demand reduction
In the first vision, AIGHP is used on a mass scale to reduce demand for reactive healthcare, primarily through more targeted and precise prevention advice.
This is a vision of the NHS in which the majority of the population would have their genome sequenced and subjected to AIGHP. The information generated would be part of a person’s medical record and would be used to provide them with a personalised risk score for various common diseases, as well as a pharmacogenomic profile setting out how they are likely to respond to common drugs and medications.
‘Every citizen would have a Personal Health Account that they control. It will store health data, including self-testing and diagnosis […] data from whole-genome sequencing identifying known risk factors including a family history of disease.’[182]
This information could be used to inform personalised lifestyle advice and prevention strategies, and to help determine a patient’s journey through the healthcare system. Individuals’ genomic risk scores could be used to help with triaging and decisions about who to prioritise for screening and diagnostic tests. They could also be used to inform prescription and treatment decisions.
‘[This data] will be transferable around the system so anywhere within the NHS or with a private provider, health data, with the consent of the patient, can be accessed.’[183]
This is a maximalist vision of AIGHP, which presumes mass participation in genomic sequencing and AIGHP, and in which the information generated by AIGHP informs every aspect of a person’s interaction with the health system.
In addition to the anticipated health benefits, it is also designed to help the NHS cope with fewer resources.
‘Healthcare demands continue to increase while costs are spiralling as health takes up an ever-higher proportion of public spending. At the same time, outcomes are deteriorating […] This calls for a paradigm shift […] we must accelerate and adopt new advances in technology that can enable health professionals to make earlier and more effective diagnoses, alongside interventions that can empower individuals to take greater personal control of and responsibility for their own health.’[184]
It is also a vision of future healthcare which, at least in some formulations, is intimately bound up with an increasingly automated, digital-first NHS. AI and automation are essential to the affordable rollout of AIGHP without overburdening the NHS workforce: This is because providing personalised healthcare advice and nudges to the whole population is expensive and time consuming unless the advice is automatically generated and delivered via digital platforms.
‘AI has the computational power to both analyse the enormity of the whole genome and generate bespoke preventative recommendations to help individuals manage their personal genetic risks.’[185]
Moreover, it is a vision that presumes a much higher degree of data sharing on the part of NHS patients and the population in general. To be accurate, predictions will need to be updated as the non-genomic data that feeds into them changes.
‘Every citizen would have a Personal Health Account that they control. It will store health data, including self-testing and diagnosis as such things become available, and from wearables like smartwatches or Fitbits.’[186]
A situational approach to AIGHP in the NHS
An alternative vision sees the technology used in a far more limited manner. Rather than viewing it as a tool with the potential to solve some of the NHS’s fundamental problems, the NHS would treat AIGHP as another emerging medical technology whose utility would need to be assessed contextually, using precautionary testing to determine its efficacy.
This is a vision in which the NHS uses AIGHP sparingly, under clinical supervision, to improve treatment and outcomes for those who are seriously ill or who have been identified as needing to take precautions regarding their health. AIGHP would be used, where recommended by a clinician, to help people understand and manage their disease risk profile, and to make predictions about how they might respond to drugs or treatments.
In some cases, the NHS might use AIGHP to inform individual health economic assessments – to determine whether it is justifiable to prescribe an extremely expensive medication to a patient, given the expected benefit.
Comparing the two visions
Many of the risks identified apply to the maximalist vision of AIGHP.
With regard to risks of surveillance and loss of privacy, the maximalist vision presupposes participation in genomic and health data sharing on a large scale and could therefore introduce structural pressures on people to share more information than they might otherwise.
The situational vision for AIGHP looks to be far more compatible with the management or avoidance of the risks identified in this report. Because AIGHP would not be used for mass prevention but would be reserved for a small section of NHS patients who needed extra health insight, there would be no expectation for most people to share their genomic and other health-relevant data.
The difference in surveillance risks also has implications for which of the two visions presents greater risks of discrimination. While neither vision is premised on genetic discrimination,[187] the structural expectations to share genomic and healthcare data that could come about with the maximalist vision of AIGHP could make it harder for a person to resist genomic discrimination (arising elsewhere) by keeping their genomic data private.
With regard to dependency, the scale of AIGHP use presumed by the maximalist vision could make a degree of outsourcing to third parties more likely. The maximalist vision also relies heavily on AIGHP insight, and automated systems more generally, and therefore looks likely to pose the risk of delegating decision making to AI systems whose reasoning is often opaque, and the possible deskilling of health professionals.
By contrast, the limited nature of AIGHP in the situational vision would mean that the NHS would be more likely to develop AIGHP insight in house, and would therefore be less likely to become dependent on a third party. Moreover, clinicians might have time to critically interact with AIGHP predictions rather than over-relying on them. This would reduce the risk of dependency on predictive tools whose reasoning may be difficult to interpret.
The maximalist vision appears more vulnerable to fragility. If AIGHP pushes the NHS towards a radically preventative mode of operation, its capacity to react to spikes in service demand would be impaired. While the promised benefits are significant, the maximalist vision for AIGHP could place significant strains on NHS resources if assumptions about its ability to reduce demand proved wrong.
Comparing the practical challenges of integrating AIGHP into the NHS, it is clear that the mass prevention vision for AIGHP would be more challenging, expensive and disruptive. If it could live up to its promises, though, it could finally shift the NHS away from a reactive, treatment-based model of health, helping people to manage upstream health risks and reduce their need for expensive health services.
Whereas the mass prevention vision would require the development and maintenance of a huge data collection, storage and processing infrastructure, more limited, clinician-mediated uses of AIGHP could likely be delivered within existing structures. And while the mass prevention model would produce far more data with which to assess the accuracy and efficacy of AIGHP, a situational model prioritising more limited, closely supervised use could potentially produce better, more contextually informed insight into how predictions were working in practice.
Conclusion
The idea that healthcare professionals might soon be able to make useful inferences about people’s future health using their DNA is no longer the preserve of horizon-scanning exercises, futures reports and science fiction. Instead, it is a possibility that the Government is actively preparing for and which many researchers, public servants and industry actors are working to realise.
While it is right to want to capture the benefits that this capability could bring, the use of AIGHP in healthcare needs to be approached with caution. The benefits brought about by AIGHP could be negated or outweighed by the harms of an insufficiently careful rollout.
We hope that this report alerts decision-makers to some of the risks of using AIGHP in healthcare, along with the potential benefits. We also hope that it sets out some of the ways in which those risks – such as loss of privacy and discrimination – can be mitigated through anticipatory changes to the regulation of data sharing in the UK.
Acknowledgements
This report was lead authored by Harry Farmer, with substantive contributions from Maili-Raven Adams and Andrew Strait.
We would like to thank all of our expert interview and public engagement participants for this project.
This project would not have been possible without the work of the members of the wider Nuffield Council on Bioethics team, including Danielle Hamm, Rebecca Mussell, Peter Mills and Catherine Joynson. We are also grateful to the members of the wider Ada team for their work on this project, including Catherine Gregory, Sohaib Malik and Octavia Reeve.
We are particularly grateful to our advisory board members who helped to steer and provide feedback on this research:
- Dr Sasha Henriques, Guy’s and St Thomas’ NHS Foundation Trust
- Laurie Smith, Nesta
- Professor Sarah Ennis, University of Southampton
- Dr Sheetal Soni, University of KwaZulu-Natal
- Dr Shwetha Ramachandrappa, Guy’s and St Thomas’ NHS Foundation Trust
- Dr Rachel Adams, Global Center on AI Governance
- Professor Joan Costa-i-Font, London School of Economics
Finally, we are grateful to the members of the Nuffield Council on Bioethics for their feedback on the final report, and in particular to Professor Sarah Cunningham-Burley.
Annex I: Addressing common arguments for keeping the ABI Code on Insurance and Genetic Testing voluntary and open ended
There are several arguments made in defence of keeping the ABI Code on Genetic Testing and Insurance voluntary and open ended. This annex sets out the most common and outlines why none of these present compelling justifications for the status quo.
Argument 1: The code needs to be open ended in case the science changes
One commonly mooted reason for keeping the code open ended is that if genomic health prediction techniques become more accurate, there may be less reason to prohibit the insurance industry from taking the insight generated into account. The code needs to be open ended, it is argued, to accommodate this possibility, and to avoid insurers being unable to deploy tools even where they meet very high standards of predictive accuracy.
Our response: This argument fails to address the fundamental concern of policymakers, academics and the public regarding the use of genomic testing in the health insurance industry. While poor predictive accuracy would undoubtedly present a compelling reason for insurers to refrain from using genomic prediction techniques, the greatest potential harms (set out in detail in the chapter on risk above) have little to do with accuracy. They relate to the possibility of those most in need of health insurance struggling to access affordable coverage. This possibility is in no way lessened by – and could well be compounded by – the emergence of increasingly accurate genomic prediction techniques. Improved accuracy, should it arise, will not diminish the need for stronger protections than currently exist.
Argument 2: It would be disproportionate to introduce new legislation given that insurers have no appetite to use genomic data to determine access to or terms of insurance
Another defence of the status quo is that because insurers have no desire and no current plans to make more extensive use of genomic data, there is no need to modify or strengthen the code. It is sometimes remarked that given the ethical and regulatory hurdles associated with the use of genomic data, integrating genomic prediction into insurance decisions is more trouble than it is worth.
Our response: Even if it is true that the insurance industry has no current desire to use genomic prediction, this has little bearing on decisions that insurers might make in the future. It is entirely possible that insurers might change their minds, especially if there are significant improvements in power, accuracy or cost.
That current choices are imperfect guides to future behaviour appears to have informed those legislatures that have introduced hard bans on the use of genetic testing in insurance. Indeed, some of the most prominent examples, such as the United States’ Genetic Information Non-Discrimination Act, are explicitly pre-emptive, not responding to current behaviour but anticipating future practices.
If there is no desire on the part of the UK insurance industry to use genomic prediction techniques, why should the hardening of the Code into law present a problem? Although there would be the costs of drafting and legislative time, this seems reasonable given the reassurance such reform would provide the public.
Argument 3: The code needs to be open ended in case an asymmetry of information emerges between insurers and potential insurance customers
Perhaps the most important argument for keeping the code open ended is the possibility that in the future, the use of genetic testing by potential insurance consumers (through direct-to-consumer genetic testing) could become common. In such circumstances, where potential insurance customers can make decisions using genomic predictions but insurers cannot, an asymmetry of information could emerge, which could be detrimental to the insurance market.
The DHSC’s 2022 three-year review of the Code on Insurance and Genetic Testing states:
‘The increased use of genetic testing may lead to a cause for concern for the insurance industry if the information individuals have about themselves – but which insurers do not ask for – changes how individuals buy insurance. If a material information asymmetry develops, whereby individual policyholders understand their risk in ways insurers are not allowed to, this could result in inaccurate pricing of insurance cover for individuals. In the longer term, this could lead to unsustainable risk management, rising insurance premiums, and reduced availability of insurance. This is why it is important for the Government and the ABI to regularly review the Code to ensure it remains relevant for both the consumer and the insurance industry.’[188]
Our response: The DHSC argues that if potential insurance customers have access to their own genomic health risk scores, those with higher risk scores will seek out health insurance at a greater rate than before, and those with lower risk scores will do so at a lower rate.
It is argued that if insurers are unable to reflect customers’ differing genomic risk scores in the premiums they offer – that is, to adjust prices at an individual level according to genomic risk scores – these insurers will have to raise the price of the average premium to reflect the higher risk level of the average consumer.[189] By pushing prices up, this would make insurance harder to access for the average consumer.
If, however, we want the insurance industry to sustainably provide universally affordable access to health insurance, it is not clear that giving insurers the ability to use individuals’ disease risk would be better than the alternative.
Instead of a rise in the average price of insurance for everyone, insurers’ use of genomic risk scores (in response to consumers’ use of genetic testing) could result in large variations in the prices of insurance for different people, with those most in need of insurance having to pay more.
As demonstrated by our public engagement research, literature review and expert engagement, this is precisely the outcome that most people and institutions want to avoid. From the perspective of fairness and universality of access, it appears worse than the cost of insurance going up for everyone.
As such, the argument in favour of keeping the code open ended (that the insurance industry may have to resort to genomic personalised pricing if access to direct-to-consumer genomic health prediction becomes widespread) could itself be seen a reason to strengthen the current ABI code into law.
Even if we assume that improved consumer knowledge of risk does justify keeping the code open ended, and allowing for future genomic personalised pricing in health insurance, it is unclear whether such a problem would emerge.
In particular, it is currently unclear whether improved access to genomic testing would result in a marked shift in consumer behaviour. A person’s genetically determined disease risk score is not the only thing affecting their approach to health insurance. People will still need protection against trauma and communicable diseases and will be inclined to seek out health insurance on that basis. Given the high stakes involved, we might also expect people to be risk averse when deciding to forgo health insurance, even in cases where they have a low disease risk score.
The argument for keeping the code open ended rests on uncertain ideas about future consumer behaviour. While there is nothing wrong with policy taking account of possible future harms, the speculative arguments about the effects of asymmetries between consumers and insurers should not be given the same weight as concrete evidence of members of the public avoiding genomic medicine and research due to worries about their data being used by insurers.
Argument 4: There is no need for a hard ban on genomic testing in insurance, because individuals can always access NHS treatment
A final argument against prohibiting the use of genomic prediction by insurers is that the UK’s health system makes it unnecessary. The suggestion is that because the NHS provides universal healthcare free at the point of need, many of the risks of the use of genomic data by insurers are significantly ameliorated. In particular, the worry that genomic personalisation could make health insurance more expensive for those most likely to need healthcare is undercut if they are able to access free NHS treatment.
Our response: There are two main problems with this argument.
First, like argument 2, this argument trades on the assumption that current conditions, in which the need for a hard ban on genomic personalisation could be argued to be weak, will obtain in the near future – and that the public trust that these conditions will remain.
While the NHS has made access to health insurance less important in the UK, this could change in the medium term.
Long waiting lists and concerns about access to care have recently led record numbers to take out private health insurance,[190] [191] with many believing that health insurance may be the only way to get decent healthcare in the future. Irrespective of whether private insurance becomes the best option for most people, it is significant that a non-trivial proportion of the population believe that it is. It also means that concerns about how genomic data might be used by health insurers cannot be assuaged by the fact that the NHS still provides a viable alternative should insurance become too expensive.
Second, it is not necessarily desirable for the NHS to become the health provider of last resort for those whose genomic risk scores make them expensive to insure. As described in the report, if health insurers are able to provide more favourable premiums to those with low genomic disease risk scores, the private sector could end up taking a greater proportion of people with low genomic disease risk, and the NHS a higher proportion of those with high genomic disease risks. This could lead to increased pressure on the NHS.
Annex II: The scenario-mapping exercise and the four futures of AIGHP in UK healthcare
Many of the conversations and interviews with experts about how AIGHP might affect the UK’s healthcare system (and broader UK society) were informed by a scenario-mapping exercise that we conducted over the course of 2023. This exercise resulted in four possible futures in which AIGHP is used in the UK healthcare systems, each of which presents a different vision for how the technology could be adopted and its impacts on healthcare and society.
In addition to indirectly informing our thinking, these four futures of AIGHP were used to inform our deliberative public engagement exercise (described in more detail in Annex III).
This section sets out in more detail the four futures of AIGHP in UK healthcare and describes the methodology we used to develop and refine them.
The four futures of AIGHP in healthcare
| First possible future The scalpel approach |
Second possible future The blanket approach |
Third possible future The prevention only approach |
Fourth possible future The automation approach |
|
| Background conditions | · high data and AI governance
· NHS monopoly · NHS budgets expand · high public trust |
· low data and AI governance
· mixed system · NHS budgets stay static / contract · low public trust |
· high data and AI governance
· mixed system · NHS budgets stay static / contract · low public Trust |
· low data and AI governance
· NHS monopoly · NHS budgets stay static / contract · low public trust |
| Summary | AIGHP is used by the NHS, under strict supervision, to supplement human care | The NHS and the private sector use AIGHP to avoid having to pay for expensive treatments | The NHS uses AIGHP to avoid having to pay for expensive treatments; face-to-face, care is only available privately | The NHS uses AIGHP to avoid having to pay for expensive treatments, but this change is less outwardly visible |
| Who develops AIGHP? | The NHS and the private sector | The NHS and the private sector | The NHS | The NHS |
| Who controls the use of AIGHP? | The NHS | The private sector | Balance between the NHS and the private sector (favouring the NHS) | Balance between the NHS and the private sector (favouring the private sector) |
| Who deploys AIGHP? | The NHS | The NHS and the private sector | The NHS | The NHS |
| What is AIGHP used for? | Better human-led, reactive care | Automated, predictive healthcare | Automated, predictive healthcare | Automated, predictive healthcare |
| Who is AIGHP used on? | A small minority of NHS patents who are ill or deemed to be at high clinical risk | Everyone | Everyone not wealthy enough to use private healthcare | Everyone |
| Where does genomic data come from? | Members of the public volunteering to share data | Members of the public sharing data as a condition of healthcare | Members of the public pressured into sharing data | International data markets and the repurposing of data shared for other purposes |
First possible future: The scalpel approach
Background conditions
The Government has turned the UK into one of the most highly regulated environments for data and AI in the world and diverts more public money to the NHS. Expanding NHS budgets allow the service to retain its de facto monopoly on healthcare provision. Relatively high regulatory and governance standards, as well as the public service ethos of the NHS, mean that most people are comfortable with the use of personal data and AI in healthcare contexts.
Who develops and controls AIGHP?
As the de facto sole provider of healthcare in the UK, the NHS has a high degree of control over the development and deployment of AIGHP. Many AIGHP products and services are developed jointly between academia, the NHS and the private sector, and the NHS contracts out some AIGHP services to private companies. However, because it holds most genomic data collected in the UK and is a huge buyer of medical services, the NHS is the senior partner in AIGHP collaborations with the private sector. The needs of the NHS largely dictate the UK research and development agenda for AIGHP, and it is in a position of power when negotiating with outside AIGHP suppliers.
What is AIGHP used for and who it is used by?
The NHS uses AIGHP sparingly, and under strict clinical supervision, to improve treatment and outcomes for the seriously ill or those who need to take particular precautions regarding their health. AIGHP is used, where recommended by a clinician, to help people understand their disease risk profile (and how it might best be managed), and to make predictions about how they might respond to drugs or treatments.
In rare cases, the NHS will use AIGHP to inform individual health economic assessments – to determine whether it is justifiable to prescribe an extremely expensive medication given the expected benefit.
The NHS does not use AIGHP to try to reduce overall demand for services. It is not offered to the whole population, or to help people better understand and cope with their disease risk profile. Along with scepticism about the efficacy of ‘personalised public health’, the NHS is not comfortable with deploying AIGHP at a scale that would make clinical oversight (and support for the subjects of the analysis) impractical.
Where does the data required for AIGHP come from?
The NHS does not require or expect patients or the population to share their genomic (or other health-relevant) data. Due to a high degree of trust in and approval of how AIGHP is used, many people share data voluntarily, either through participation in studies or (more commonly) by agreeing for data collected through treatment to be used for research. Genomic data is stored by the NHS on a federated data platform, such as a secure data environment.
For people with rare and ultra-rare diseases, genomic data is still stored securely but the NHS relies on agreements with other health services to contribute to and access international aggregate datasets.
What is the role of direct-to-consumer genetic testing?
Direct-to-consumer genetic testing is available, but the NHS generally refuses to let such tests inform treatment decisions due to scepticism as to their reliability. They are used mainly by the rich for personal interest or to inform the decisions of the health conscious.
Second possible future: The blanket approach
Background conditions
The Government has pursued a deregulatory agenda for data and AI. Contracting NHS budgets mean more people jumping ship to the private sector, which has expanded its share of the UK’s healthcare provision, and the UK now has a mixed system of healthcare. The NHS is one of many healthcare providers, alongside a large and growing ecosystem of private healthcare companies, which are generally funded by insurance. Low levels of regulation and the involvement of for-profit companies (as well as the increasing relevance of health insurance) have made the public wary about sharing sensitive data for healthcare purposes.
Who develops and controls AIGHP?
Most AIGHP products and services are delivered and controlled by the private sector, which has bought a lot of the necessary data, intellectual property and expertise from the NHS, universities and elsewhere. The NHS has shared much of its historical aggregate dataset with private companies in exchange for some degree of access to AIGHP services. Internal commercial industry committees determine some of the standards for the bigger predictive models and are responsible for organising a degree of data sharing across an otherwise fragmented data ecosystem.
What is AIGHP used for and who it is used by?
AIGHP is marketed at and available to the majority of the population – not just those who are ill or at a high risk of serious health problems.
Within the NHS, AIGHP has been bought in to more efficiently allocate smaller real-terms budgets and preserve the time of an overstretched workforce.
The NHS uses genomic data about population-level variations in disease risk to inform commissioning decisions and to more efficiently allocate resources. The decision to pay for a prescription is often guided by pharmacogenomic insight, with drugs that show considerable genomic variation in efficacy prescribed only to those who it is predicted will show a sufficiently positive response.
It also uses AIGHP for far more targeted public health interventions than would otherwise be possible. NHS patients who share their genomic data are given personalised information about their genomically determined disease risk profile, so they can make personal lifestyle and other behavioural changes to minimise risk and nudges can be sent to their phones.
To cope with a shrinking workforce, the NHS has invested in AI chatbots to replace the role of most medical receptionists and some diagnostic and therapeutic interventions, and as gateways to more specialised care. The data from wearables and genomic screening is used to compensate for the loss of rich contextual information gained from face-to-face contact with a medical professional. These AI systems can look at people’s wearable data, self-reported symptoms and AIGHP-generated predictions to make reasonably accurate assessments about where best to refer people or what drugs or medicines to prescribe. In cases of low certainty they may refer patients to a human clinician, but many referrals and prescriptions can be generated directly through the NHS app.
The business model of all but the most expensive private healthcare providers also focuses on prevention and the precise allocation of scarce, expensive healthcare resources. Similarly to the NHS, the bulk of what most private healthcare companies provide is health tracking, genomically tailored AI-generated advice and sometimes the use of this insight for prescribing. Private healthcare providers may sometimes provide wearables and other tracking technologies to support the use of AIGHP, to provide more tailored / personalised behavioural nudges than those available in the NHS.
In addition to using AIGHP to allocate resources, and to provide personalised behavioural nudges to customers, a key use of AIGHP by the private healthcare sector is in determining the terms of health insurance. Most health insurance packages require people to share their genomic data, to determine their insurance risks and therefore the cost and terms of their insurance. It is common for private healthcare providers to make insurance coverage for those deemed to be poor genomic health risks contingent on lifestyle changes to mitigate such risks.
Where does the data required for AIGHP come from?
Because of low public trust in the use of data and AI in healthcare (and awareness of how this data is used to determine insurance rates), few people voluntarily share their genomic data for medical and scientific research. Sharing genomic data becomes a de facto requirement of interacting with most healthcare services. The NHS is explicit that you will receive an inferior standard of care if you do not share your genomic data, and genomic data sharing is a requirement of all but the most expensive private health insurance.
To improve the representativeness of their respective genomic datasets, the NHS and the private sector are engaged in (and constantly renegotiating) several data-sharing agreements. Low data protection regulation means that genomic data is poorly protected.
What is the role of direct-to-consumer genetic testing?
Direct-to-consumer genetic testing is common. One of its biggest and most controversial uses is where individuals try to determine if it is worth getting private health insurance. Though of highly variable accuracy, direct-to-consumer genetic tests inform many decisions not to get private health insurance – thereby depriving insurers of some of their most lucrative customers. As a result, many healthcare companies have attempted to buy up and close down direct-to-consumer services in a so far unsuccessful attempt to stop this practice.
Third possible future: The prevention only approach
Background conditions
The Government has been forced to keep relatively high data and AI regulation standards, partly as a way of winning over a wary public but mainly out of a need to retain parity with other parts of the world.
At the same time, it has reduced health budgets in real terms, which has led to greater prominence of private healthcare providers in the UK’s healthcare mix. Long waiting times and declining standards in the NHS, plus the profit motive of private companies (and the increasing relevance of health insurance), have made the public distrustful of the use of data and AI in healthcare, despite relatively strict regulation.
Who develops and controls AIGHP?
Regulatory constraints on the collection and sharing of personal data have made it hard for many private sector entities to collect and collate genomic data. Research and development of AIGHP tools is done by collaboration between the NHS and the private sector, with the former providing the data and the latter providing a large proportion of the funding and the computing power and expertise.
What is AIGHP used for and who it is used by?
AIGHP is far more extensively used in the NHS than in the private sector.
As with the second possible world, the NHS uses AIGHP to inform service-wide commissioning decisions. It also uses it to provide individuals with targeted public health advice: patients who share their genomic data are given personalised information about their genomically determined disease risk profile and by default have behavioural nudges sent to their phones, tailored to their data.
As a result of AI regulation (and specifically, requirements around the accountability of automated decision-making systems), the NHS has been hesitant to use AIGHP to ease longstanding labour shortages. AI-powered systems (such as chatbots) using genomic insight to determine where to refer patients or to make prescription decisions are rare, and genomic predictions are used to guide clinical decision making only on an ad hoc basis, with the oversight of clinicians.
Due to the costs associated with regulatory compliance and difficulty of collecting sufficiently extensive datasets, private healthcare providers do not make extensive use of AIGHP. Instead, private sector health provision is very expensive and provides something much closer to a traditional, reactive model of care.
Where does the data required for AIGHP come from?
Regulatory constraints on the collection and sharing of personal data have made it hard for many private sector entities to amass enough genomic data to develop viable AIGHP tools.
In the UK, most genomic data used for AIGHP training is collected by the NHS – which is large enough, better configured and more incentivised to cope with regulatory compliance, and which holds this data in a ‘secure research environment’.
While genomic data sharing is not a condition of receiving NHS treatment and the right of patients to withhold such data is formally respected, narratives around ‘genomic data solidarity’ and ‘genomic data free-riding’ – which hold that everyone has to do their part by sharing their data with the NHS, and that those who fail to do so despite benefiting from genomic predictions are selfish – are tolerated, if not actively propagated, by the NHS and the Government. As a result, many of those who use the NHS agree to share their genomic data with the service.
What is the role of direct-to-consumer genetic testing?
Direct-to-consumer genetic testing has a niche market, and is most often used by the wealthy, who want additional insight to supplement the traditional, expensive healthcare they receive privately.
Fourth possible future: The automation approach
Background conditions
There have been no big changes from what we have now. The Government has not introduced any new data or AI regulation, so the UK has failed to keep pace with technological advances and has become permissive by international standards. Despite falling real-terms budgets, the NHS has held onto its de facto monopoly over service provision – partly because the private sector has been slow to fill the gap and partly because there is limited demand for private healthcare due to the weak economic outlook.
Poor NHS provision and low levels of data regulation mean that the public are generally reluctant to share data for health purposes and unhappy with AI being integrated into health – which they see as a poor substitute for human care and judgement.
Who develops and controls AIGHP?
As the sole de facto provider of healthcare in the UK, the NHS is the main deployer of AIGHP, but it plays a less prominent role in developing AIGHP tools.
The NHS trades genomic data for AIGHP tools. However, because lower regulatory standards make it easier for private companies to get genomic data directly from the UK population (or from third parties), the NHS receives worse terms than in other futures with more stringent regulation – getting less functionality in exchange for more data, and having less of a say over the data it receives. The NHS is under intense pressure to ensure that AIGHP tools result in cost savings.
What is AIGHP used for and who it is used by?
The NHS is by far the biggest deployer of AIGHP tools in the UK and makes extensive use of them to cope with limited budgets and political and public pressure to improve service quality.
AIGHP is used on practically everybody who interacts with the NHS. However, due to low public trust in the use of data and AI in healthcare, and particular resentment towards AIGHP (which is seen as invasive and inferior to human-driven, reactive care), its use is rarely as overt as in other possible futures.
The NHS uses genomic data about population-level variations in disease risk to inform commissioning decisions and to more efficiently allocate resources. The decision to pay for a prescription will often be guided by pharmacogenomic insight, with drugs that show considerable genomic variation in efficacy prescribed only to those who it is predicted will show a sufficiently positive response.
Unlike in other possible futures, the NHS does not use AIGHP to provide service users with insight into their disease risk profiles and does not provide healthcare nudges.
The NHS uses AIGHP as a means of coping with a small, overstretched workforce, but this is less visible from the outside. Rather than being used to more effectively automate referral and triaging services, AIGHP is used for clinical decision support, enabling clinicians to deal with a larger number of patients and (theoretically) reducing the need to have more senior consulting staff on call. This practice has led to serious concerns about the long-term deskilling of the NHS, as the use of these systems lets it get away with training and retaining fewer senior doctors.
Where does the data required for AIGHP come from?
Due to low public trust in the use of data and AI in healthcare, the NHS struggles to rely on the UK population voluntarily sharing genomic data to train AIGHP systems. In response, the NHS loosens restrictions on the repurposing of medical data, to generate bigger datasets.
What is the role of direct-to-consumer genetic testing?
Direct-to-consumer genetic testing is used by people to better understand (and occasionally challenge) NHS treatment decisions. Due to low levels of regulation, there are large variations in the predictive accuracy of such services.
Methodology
These four futures of AIGHP in healthcare were developed in collaboration with our advisory board and a panel of external experts using a variant of a technique known as ‘morphological analysis’. Our approach had the following broad components:
- We identified a topic whose potential futures were to be explored (in this case, the potential futures brought about by AIGHP) and specified the geographical and temporal parameters for that exploration (in this case, the UK within the next 5–10 years).
- We worked with our advisory board and expert panel to identify four ‘critical uncertainties’ – clearly defined aspects of the future that are uncertain and will affect how the topic in question might develop.
- Using these four ‘critical uncertainties’ we plotted 16 possible futures, each based on different permutations of the four critical uncertainties.
- We identified the four most credible and distinctive possible futures from the initial 16.
- We held two workshops to elaborate on the features of these four possible futures, to think in a structured manner about the pressures and dynamics that AIGHP might create in different circumstances.
Identifying critical uncertainties
Critical uncertainties are factors that both:
- Are capable of significantly impacting the future of the topic in question.
- Exhibit a particularly high degree of uncertainty, with little consensus or confidence among experts regarding likely outcomes.
In collaboration with our advisory board and external experts, we selected four critical uncertainties:
High versus low data and AI governance
| High data and AI governance standards | Low data and AI governance standards |
| The UK adopts more stringent governance regulatory standards on:
· how companies and the public sector collect, store and process personal data, including genomic data · the training and delegation of decision making to AI systems. |
The UK takes the decision to radically diverge from the current, EU-influenced model of data protection and take a laisse faire approach to AI regulation. By the standards of rich countries, the UK places few constraints on data collection, storage and processing, or on the development and deployment of AI systems. |
Public versus mixed healthcare provision
| NHS monopoly | Mixed system |
| The NHS continues to be the sole de facto provider of medical services in the UK, even if large parts of it are ‘marketised’. While the NHS may continue to struggle, NHS funding is still a big political issue because practically everyone depends on it, and the availability of state-of-the-art treatments on the service is considered important. | Falling standards and long NHS waiting times mean more and more people seek treatment privately, causing a surge of investment in private healthcare in the UK, growing the size of the sector considerably. The more people jump ship for the private sector, the faster this shift becomes, as middle-class support for the service is eroded and more NHS workers move over to the private sector. |
Expanding versus contracting (real-terms) NHS budgets
| NHS budgets expand | NHS budgets stay static / contract |
| In response to falling standards and workforce retention issues, the Government takes the decision to dramatically increase UK healthcare spending, bringing per-person spending up to a level commensurate with comparable European countries. | The Government refuses to devote substantial extra money to the NHS, keeping funding steady but failing to account for rising demand and increasing costs. |
High versus low public trust of data and AI use in healthcare
| High public trust | Low public trust |
| The public are generally supportive of the use of data and AI in healthcare contexts, and are, for the most part, willing to share data for healthcare purposes and to allow AI to inform clinical decision making. | The public are generally wary of the use of data and AI in healthcare contexts, with a significant proportion actively hostile. As a result, many people try to avoid sharing healthcare data when not absolutely necessary, and the use of AI in medical settings is subject to significant public and media scrutiny. |
Plotting out the 16 possible futures
Once the four critical uncertainties were identified and defined, producing the 16 resulting possible futures (PFs) was simply a matter of plotting out their different permutations. The table below illustrates how this can be done if each critical uncertainty is expressed as two possibilities (e.g., ‘high A’ versus ‘low A’).
| High A | Low A | ||||
| High B | Low B | High B | Low B | ||
| High C | High D | PF 1 | PF 2 | PF 3 | PF 4 |
| Low D | PF 5 | PF 6 | PF 7 | PF 8 | |
| Low C | High D | PF 9 | PF 10 | PF 11 | PF 12 |
| Low D | PF 13 | PF 14 | PF 15 | PF 16 | |
Describing the final four possible futures
We used two workshops of external, expert stakeholders and members of our advisory board to help us describe the final four possible futures in greater detail.
To produce a useful set of outputs, capable of illustrating and informing discussion of the ways in which AIGHP might impact on people and society, we aimed for each description to address the following aspects of its given future. These parameters were given to the workshop participants in advance, and provided as prompts throughout the workshops.
Who produces, controls and has access to AIGHP capacity:
- which groups and actors hold AIGHP capacity
- where the data and other necessary inputs for AIGHP comes from, how it is collected and how access to it is managed
- who has (formal and de facto) control over the use of AIGHP and the basis of this control
- who has access to AIGHP, and the terms of that access.
The forms taken by AIGHP:
- the kinds of business models and products that are built on top of AIGHP systems and capabilities
- how the uses and capabilities of AIGHP are affected (limited, constrained or directed) by policy, law, regulation, norms and economic factors
- how other actors, such as individuals, private companies and non-health-related public bodies, make use of AIGHP – if at all.
How healthcare and public health systems are structured and delivered, given the availability of AIGHP:
- how healthcare and public systems make use of AIGHP, in what circumstances and to what ends
- what healthcare and public health systems provide to members of the public, and what they expect or require from members of the public
- the level of demand for AIGHP on the part of healthcare and public health systems.
Finally, the staff team wrote up ‘pen sketches’ of each of the four possible futures, in accordance with the descriptions given in the workshops. For the sake of completeness, at some points the staff team elaborated on details that were covered in the workshops.
Annex III: The deliberative public engagement exercise
Aims and methodology
As part of AI and Genomics Futures, the Ada Lovelace Institute and the Nuffield Council on Bioethics conducted a deliberative public engagement exercise to understand public views and priorities on the governance, regulation and cultivation of AIGHP. This took place between mid-August and late September 2023 and was conducted by Thinks Insight.
Twenty-four members of the English public were involved. Twelve participants were recruited from Manchester and surrounding areas and 12 were recruited from Peterborough and surrounding areas. To ensure each group was broadly reflective of the population in England, the recruitment involved quotas based on a set of socio-demographic strata agreed between the AI and Genomics Future team and Thinks Insight. These strata included characteristics such as gender, age, location, education, and living with a disability or long-term health condition, among others (see below for the recruitment sample). Due to cost restrictions, we were unable to recruit from Wales, Northern Ireland or Scotland. This highlights a limitation of our method – our public engagement feedback is not representative of members of the public from all parts of the UK. Findings below that reference the impacts of AIGHP on the UK healthcare system refer to participants considering how AIGHP would impact the health systems of England and the devolved nations.
The engagement process consisted of:
- An online community, which was an opportunity for participants to familiarise themselves with the subject matter and the process they were going to be taken through for the rest of the engagement.
- Two day-long in-person workshops, one for the Manchester group and one for the Peterborough group. The aim was to understand and discuss AIGHP and the ways it might be deployed, given different scenarios of the future of healthcare in the UK. Participants were presented with four possible futures of AIGHP, intended to illustrate some of the ways the technology might affect people and society. These scenarios were informed by: (1) a horizon-scanning exercise that involved 15 stakeholders from academia and from the private and public sectors; (2) a scenario-mapping exercise that involved ten stakeholders and input from the advisory board of the AI and Genomics Futures.
- Two online deliberative sessions attended by the whole group of 24. In the first online session, participants were given presentations by two expert witnesses (Sarah Henriques, a genetic councillor from Guy’s and St Thomas’ Foundation Trust, and Maxine Mackintosh, Programme Lead for diverse data at Genomics England). Participants were able to ask questions about the subject matter to inform their views and discussions. In the second online session, participants were given the opportunity to refine potential recommendations for what they believed should be done to enhance the benefits and mitigate the harms and risks of AIGHP.
The four futures of AIGHP – and the context of participants’ views
Participants were introduced to the concept of AIGHP and were presented with four scenarios illustrating very different ways that the capability could be incorporated into the health service. The aim of presenting these four scenarios was to illustrate how many of the advantages and risks of AIGHP were contingent on the different systems in which they were used.
While participants did have strong, clearly articulated views on the pros and cons of the four different possible futures, the purpose of the scenarios was to draw out participants’ more general views, priorities and values when it came to AIGHP. Participants were asked not which (if any) of the possible worlds they would most or least like to become a reality but instead to reflect on what risks would concern them, what they would like to see decision-makers do to guard against them and which benefits they would like to see decision-makers actively pursue.
As such, we have tried to show which comments, views and opinions were specific reactions to the circumstances described in particular possible worlds and which were more general comments on the nature of (and people’s aspirations for) AIGHP.
Hopes and positive sentiments
Over the course of the engagement exercise, participants were positive about the benefits AIGHP could bring to healthcare.
At various points, participants approved what they saw as the potential of AIGHP to enable faster, more accurate treatment decisions, cut costs and help enable a transition towards a more preventative model of healthcare. Specifically, participants spoke about the ability of AIGHP to enable:
- More targeted treatments: Some participants commented positively on the ability of AIGHP to shorten the process of finding appropriate treatments for patients with specific needs.
- Better informed clinical decision making: Some participants were enthusiastic about the idea that AIGHP could provide individuals and clinicians with more information to better diagnose and treat illnesses.
- A more predictive model of healthcare: Participants were also broadly (though not unconditionally) positive about the perceived ability of AIGHP to support a more preventative approach to healthcare. (Further discussion of this point is covered below.)
Participants’ belief in the potential benefits of AIGHP was supported by the commonly expressed view that access to AIGHP insight should be as widely and fairly distributed as possible. This was most clearly demonstrated by participants’ objection to the first possible future (in which the NHS offers AIGHP only in extreme cases) on the grounds that relatively few people had access to AIGHP. Issues of equality in access were therefore another important theme, which we cover further below under the section on concerns.
Other positive views regarding AIGHP were more heavily caveated. Notably, several participants suggested that AIGHP may simply be the least bad option for the UK health service, given tight budgets and high demand. These participants thought that even though AIGHP would come with drawbacks, these may be worth tolerating if the technology can help to address current problems in UK healthcare.[192]
It was striking that throughout the dialogue, participants were generally positive about AIGHP in the abstract but expressed greater scepticism when invited to consider how the technology might be deployed. The framing of the dialogue, with its emphasis on participants thinking about what they would need to feel comfortable with AIGHP, could have pushed participants in a more negative direction.
Concerns
A broad sentiment expressed by participants was that AIGHP was a technology with significant prospective benefits but potentially catastrophic harms should it be mismanaged or should things go wrong. While participants did not explicitly state which harms and risks of AIGHP they considered to be catastrophic, it was clear that many regarded AIGHP as a highly risky technology, the use of which would need to be carefully regulated and monitored.
Some of the most prominent worries and concerns around AIGHP expressed by participants are described below.
Personal data collection, use and storage
The need for collection, storage and processing of genomic data (along with the phenotype data required for genomic analysis) was a point of contention for participants. While most accepted that genomic data collection was a prerequisite of conducting genomic research and analysis, views differed on the terms and circumstances under which data could and should be collected.
Participants were concerned about potential threats to privacy arising from having to share genomic or phenotype data, noting that many people would likely be resistant to requirements to share such personal, sensitive information.
More specifically, participants voiced concerns that data could be stored for longer than strictly required or used for purposes other than those specified, that it might be used in a manner contrary to the interests of those sharing it, and that it might be accessible by third parties.
‘So what’s to stop them saying “Oh, we’re going to give that data to the bank” who say: “Actually, we’re not going to give you a mortgage, because, you know, you might die in 10 years”. Like it could affect everything, could affect all aspects of your life.’
Participants also expressed a desire to retain control over genomic data that they share, and to be able to access their genomic information and interpret it themselves.
Participants were also sensitive to the need to obtain consent for genomic data sharing and processing, and the trade-offs and tensions that can arise in assuring that consent for the collection and use of genomic data is adhered to, is meaningful, and does not impede important research or medical uses. Specifically, views differed on how the need to collect data only with consent could be reconciled with the need for a large proportion of the population to share their genomic data to make AIGHP work.
This tension around consent also arose in conversations about whether people should be required or incentivised to share genomic data to ensure that genomic datasets are large and diverse enough to produce robust, accurate insights.
Participants were generally more comfortable with the NHS having genomic and phenotype data than the private sector. Many of the concerns around data processing and storage revolved around this data leaving the NHS and finding its way into the hands of third parties, and particularly the private sector.
Cost and opportunity cost
Participants also raised questions about the cost of developing and maintaining AIGHP systems, particularly given current resource constraints on healthcare: while some thought the costs of AIGHP were acceptable given the expected cost savings, others questioned whether AIGHP should be a priority given the NHS’s current failure to deliver basic services. The development and rollout of AIGHP could also constitute a significant investment in a technology before its value was fully known.
‘Is this going to take such a long time for all of this data to be collected? It seems it’s going to go on and on for years and year and cost a lot of money. What about people that need to know things now? And will it go anywhere in the end, will it be successful with all this technology that’s going into it?’
Accuracy and reliability of AIGHP predictions, given data and system limitations
Questions about the accuracy and reliability of AIGHP systems were raised at various points throughout the dialogues. Notably, participants were conscious that the accuracy of AIGHP would be contingent on the accuracy, quality and diversity of training data and the quality and accuracy of the information provided by the end user.
There was also a general reluctance to entrust something as important as health to automated systems whose workings and reasonings can be obscure and difficult to assess and audit.
Ability of AIGHP systems to cope with the complex, messy reality of healthcare
Another source of poor accuracy and reliability suggested by participants was the poor capacity of AIGHP to cope with the messy, human components of healthcare provision and decision making. A degree of scepticism was expressed regarding the ability of AIGHP systems to reliably contextualise data that they collect from health service users. In particular, participants mooted the possibility that systems might misinterpret data, fail to consider contextual cues or be fed data that was, by accident or by design, misleading.
Related to these concerns were suggestions that many people would be reluctant to follow health advice or ‘nudges’ generated by an AI system, regardless of whether such information was informed by genomic (and other health) data. People with poor digital literacy, or with limited access to digital services and devices, would present a major challenge for the rollout of some AIGHP services, and the impersonal nature of these systems would put many people off using them.
In response to these concerns, many participants wanted the accuracy and reliability of AIGHP systems to be tested and proven at a small scale before the technology was rolled out more widely, with some mentioning the need for pilots.
Insurance and how the private sector might use AIGHP
A common concern expressed by participants (in response to the four futures of AIGHP, but also in advance of being presented with these scenarios) was the use of AIGHP-generated insight about individual or group disease risk by insurance companies. Participants expressed unease at the prospect that genomic health predictions might be used to deny some people access to health insurance, or to offer those deemed to have higher disease risk more expensive insurance premiums.
Participants were also generally concerned that insurance companies might offer those at higher risk of certain diseases insurance premiums only on the condition that they demonstrably take steps to reduce their disease risk. This was generally thought to be unfair, to place undue stress and burden on those affected, and to risk compounding existing health inequalities.
‘My dad, for example, he’s diabetic, it’s type two. He doesn’t follow the diet tips that they send him. He doesn’t listen to anything that’s told to him. He’s one of the older generation. With insurance premiums in this scenario: if he wasn’t recording that he was following his lifestyle tips, his premiums would go up and he wouldn’t be able to afford it. He is working class. It sounds like we’d be held hostage.’
There was also concern about the use of AIGHP insight by insurers in circumstances where private healthcare exists alongside NHS provision. Specifically, some participants worried that if private insurers were to use AIGHP to deny insurance to or raise the price of premiums for those with high risk, this would lead to the NHS having to take on all the sickest, most expensive people, putting further strain on the service.
Discrimination and eugenics
Alongside concerns about the use of genomic data and AIGHP insight by insurance companies, many participants expressed broader worries about the potential of AIGHP to enable genomic or genetic discrimination. Others went further, venturing that the availability of predictive insight into people’s disease risk might, in the long run, encourage or enable eugenics.
‘I worry it’s going to be a two-tiered system, the haves and have nots, when you choose to keep the stronger richer people alive.’
Inequality (of access and of outcomes)
A common, overarching set of concerns raised by participants was the potential for AIGHP to generate new and exacerbate existing inequalities. In addition to concerns about insurance and genetic discrimination, participants were concerned about the possibility of access to AIGHP being confined to some groups, such as the rich, the young and the digitally literate, and denied to others, i.e. the poor, the elderly and those unable to use digital technologies. Participants also expressed concern that AIGHP predictions might work better, or be more accurate or reliable, for individuals with certain genetic ancestry or demographic groups than for others, leading to a divergence of healthcare outcomes for different groups.
Automation and the division of labour between AI and humans
Another common set of concerns was around the desirable division of labour between AIGHP systems and human labour and expertise.
Many participants objected to the idea (set out in some of the stimulus materials) of using AIGHP to inform automated triaging and GP services, on the grounds that they could fail and would only be as good as the data fed into them.
A notable counterpoint, albeit voiced by a minority of participants, was that such forms of automation may be preferable to the current situation with the NHS, as they would facilitate faster and more universal access to basic medical advice and to referral within the system.
Many participants expressed worries about health systems coming to over-rely on AIGHP in clinical decision making. Here, concerns revolved around the potential for AIGHP to make mistakes, as well as the more structural risk that its availability could lead health systems to train and hire fewer human professionals. It was noted that this could leave health systems especially exposed in the event of a failure of AIGHP systems. For this reason, many participants expressed the preference that AIGHP be used only with strict human supervision, and in a supporting role to human decision making and expertise.
‘You’re going to get less qualified doctors, so if it all crashes then you’re going to end up with less qualified people looking after you.’
A radically preventative model of healthcare
A final set of concerns, voiced by a smaller proportion of participants than the above worries and more contentious among the group, concerned the radically preventative model of healthcare that AIGHP could enable.
Some participants were concerned that the use of AIGHP to shift the UK health systems towards a focus on prevention could place unfair and unrealistic expectations on individuals to keep themselves healthy. Some participants suggested that the ability of AIGHP to provide individuals with genomically personalised insight and health advice would in many cases be unlikely to enable them to significantly affect their chances of getting ill. Participants said that in many cases, broader environmental determinants of health and lifestyle would make it difficult for people to act on such advice. Some participants argued that a shift towards prevention could leave many feeling exposed in the event of developing a serious health condition.
In contrast, many participants felt that the shifting of responsibility for maintaining health onto individuals was a selling point.
‘It’s great in a perfect world, but it’s fundamentally at odds with what the NHS currently does, providing free care to everybody, irrespective of if you’ve done something really stupid, or you make terrible lifestyle choices, you know, you might, but you still get the same treatment.’
Overall conditions for AIGHP being used and thoughts about how it should be used
Over the course of the dialogue, participants consistently expressed several desires and expectations for the use of AIGHP.
Regulation and oversight
Participants’ support for the use of AIGHP, and for the collection and processing of genomic data, was strongly contingent on strict regulation. Throughout the dialogue, calls for regulation of AIGHP were consistent. Though accounts differed as to what this regulation would cover, some common themes were:
- regulation and norms to mandate consent, transparency and clear communication around the collection, processing and retention of genomic data and generated insight
- limitations on the use of genomic data by insurance companies
- measures to ensure the accuracy and reliability of AIGHP, and to mitigate against the failure of AIGHP systems.
Communication, education and public awareness
Participants consistently called for transparency and clear communication regarding when and to what end genomic data is collected, and that any genomic data programmes need to be clearly communicated with those directly involved and with the public. In many cases, clear communication was seen as a prerequisite for meaningful consent to genomic data sharing and processing. Participants also stated that given the complex, technical nature of genomic science, AI and AIGHP, communication would need to be complemented by broader public education and awareness programmes so the public are better equipped to understand the reasons for and implications of sharing their data.
Retention of human agency over medical decisions (AI should not replace doctors) and avoiding over-reliance on AI systems
Another common concern was the possibility that AIGHP would lead to deskilling of the medical profession. There was also a worry that reliance on AIGHP systems might disempower patients and clinicians. The principal worry was about AIGHP systems failing, which could result in poor decisions unchecked by humans and in the NHS being unable to cope, having become over-reliant on these systems.
‘These days you can get an airplane to take off and land anywhere automatically, literally, but you wouldn’t get on it if there weren’t any pilots, would you?’
Desire for universal access to AIGHP insight
A notable point of agreement was that ideally, everybody should have access to AIGHP oversight rather than just a select few. There were strong fears that AIGHP might end up the preserve of the rich. A caveat is that views regarding the first possible world, in which AIGHP is limited to the very sick, softened when the reasons for this restriction in access were set out in more detail.
‘Everyone gets access to it. That’s a positive. Unlike in the previous scenario.’
Notable points of disagreement
The terms on which genomic data should be collected
A majority of participants stressed the importance of genomic data being collected only with the meaningful consent of subjects, and that people should not be put under pressure to share their genomic data. However, a significant minority expressed worries that without a degree of pressure or incentivisation, not enough people would share their genomic data to make the system viable.
The role of the private sector
There was general unease about involving the private sector in AIGHP and a general preference for genomic data and processing to stay in the NHS. This was counterbalanced by some suggestions that the private sector was better placed to innovate in AIGHP and manage the technology. Moreover, a minority of participants expressed concerns that overly strict regulation, or stopping the private sector getting involved in AIGHP altogether, might hamper innovation and slow down the rollout of AIGHP services. Many people thought that AIGHP provided by the private sector would be better than no access to AIGHP at all.
About the Nuffield Council on Bioethics
Developments in biomedicine and health are essential to solving the world’s problems but can also raise profound ethical challenges. The Nuffield Council on Bioethics (NCOB) was established by the Nuffield Foundation in 1991 to help address those challenges and ensure changes in biomedicine and health benefit everyone equitably and fairly. Since 1994, we have been co-funded by the Nuffield Foundation, Wellcome and the Medical Research Council.
The NCOB is a leading independent policy and research centre, and the foremost bioethics body in the UK. We are made up of a team of Council members and Executive staff who identify, analyse and advise on ethical issues in biomedicine and health so that decisions in these areas benefit people and society.
Through our horizon-scanning programme, we monitor bioscientific and medical developments that raise ethical questions and could have impacts on society. We aim to anticipate these developments at an early stage, so that we can consider them and make appropriate recommendations in a timely way.
For over 30 years, we have identified and tackled some of the most complex and controversial issues facing societies across the globe. We have brought clarity to complexity and plotted practical ways through seemingly intractable dilemmas. This has led to shifts in public understanding and lasting policy change in the UK and internationally.
Find out more:
Website: nuffieldbioethics.org
Email: bioethics@nuffieldbioethics.org
Footnotes
[1] UK Research and Innovation, ‘Accelerating Detection of Disease’ (May 2023) <www.ukri.org/what-we-do/browse-our-areas-of-investment-and-support/accelerating-detection-of-disease> accessed 4 June 2024.
[2] Our Future Health, ‘The UK’s Largest Ever Health Research Programme to Transform the Prevention, Detection and Treatment of Diseases’ (January 2022) <https://ourfuturehealth.org.uk/news/the-uks-largest-ever-health-research-programme-to-transform-the-prevention-detection-and-treatment-of-diseases> accessed 24 April 2024.
[3] Throughout this report, unless otherwise stated, ‘the Government’ refers to the UK Government in Westminster, as opposed to the governments of the devolved administrations. ‘The NHS’ refers to the four devolved health systems in the UK – NHS England, NHS Wales, NHS Scotland and NHS Northern Ireland.
[4] Office for Life Sciences and others, ‘Genome UK: The Future of Healthcare’ (September 2020) <https://assets.publishing.service.gov.uk/media/5f6b06a9d3bf7f723ad68ccc/Genome_UK_-_the_future_of_healthcare.pdf> accessed 2 August 2024.
[5] NHS England, ‘Accelerating Genomic Medicine in the NHS’ (October 2022) <www.england.nhs.uk/publication/accelerating-genomic-medicine-in-the-nhs> accessed 24 April 2024.
[6] Ada Lovelace Institute, ‘AI and genomics futures’ <www.adalovelaceinstitute.org/project/ai-genomics-futures> accessed 2 August 2024.
[7] Harry Farmer, DNA.I.: Early Findings and Emerging Questions on the Use of AI in Genomics (Ada Lovelace Institute and Nuffield Council on Bioethics, August 2023) <www.adalovelaceinstitute.org/wp-content/uploads/2023/08/Ada-Lovelace-Institute-NCOB-DNAI-genomics.pdf> accessed 2 August 2024.
[8] Richard M Turner and others, ‘Pharmacogenomics in the National Health Service: Opportunities and Challenges’ (2020) 21 (17), 1237 Pharmacogenomics <https://doi.org/10.2217/pgs-2020-0091> accessed 2 August 2024.
[9] Anne Gulland, ‘UK Has Best Health System in Developed World, US Analysis Concludes’ (2017) 358, j3442 BMJ <https://doi.org/10.1136/bmj.j3442> accessed 2 August 2024.
[10] ‘Staff Shortages’ (The King’s Fund, 2024) <www.kingsfund.org.uk/insight-and-analysis/data-and-charts/staff-shortages> accessed 23 April 2024.
[11] Office for National Statistics (ONS), ‘NHS Waiting Times, 16 January to 15 February 2024 – Office for National Statistics’ (April 2024) <www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/healthcaresystem/datasets/nhswaitingtimes16januaryto15february2024> accessed 18 June 2024.
[12] Michael Searles, ‘NHS Waiting List Could Be Two Million Longer Than Thought’ Telegraph (3 April 2024) <www.telegraph.co.uk/news/2024/04/03/nhs-waiting-list-could-be-two-million-longer-ons> accessed 3 August 2024.
[13] ‘Public Attitudes to the NHS and Social Care’ (National Centre for Social Research, 12 June 2024) <https://natcen.ac.uk/publications/public-attitudes-nhs-and-social-care> accessed 3 August 2024.
[14] Denis Campbell, ‘Public Satisfaction with the NHS at Its Lowest Ever Level, Poll Shows’ Guardian (27 March 2024) <www.theguardian.com/society/2024/mar/27/public-satisfaction-with-the-nhs-at-its-lowest-ever-level-poll-shows> accessed 3 August 2024.
[15] Alvin Powell, ‘Risks and Benefits of an AI Revolution in Medicine’ Harvard Gazette (11 November 2020) <https://news.harvard.edu/gazette/story/2020/11/risks-and-benefits-of-an-ai-revolution-in-medicine> accessed 3 August 2024.
[16] David Wallace-Wells, ‘Suddenly, It Looks Like We’re in a Golden Age for Medicine’ The New York Times (23 June 2023) <www.nytimes.com/2023/06/23/magazine/golden-age-medicine-biomedical-innovation.html> accessed 3 August 2024.
[17] Jon Heggie, ‘Genomics: A Revolution in Health Care?’ Science (20 February 2019) <www.nationalgeographic.com/science/article/partner-content-genomics-health-care> accessed 3 August 2024.
[18] Chris Ham, ‘The Rise And Decline Of The NHS In England 2000-20’ (The King’s Fund, April 2023) < https://assets.kingsfund.org.uk/f/256914/x/0ab966500b/rise_decline_nhs_england_2000-20_2023.pdf> accessed 2 August 2024.
[19] Nicola Blythe and Shilpa Ross, ‘Strategies to Reduce Waiting Times for Elective Care’ (The King’s Fund, December 2022) <https://assets.kingsfund.org.uk/f/256914/x/43ac620e93/strategies_reduce_waiting_times_2022.pdf> accessed 2 August 2024.
[20] ‘UK Health Spending over Past Decade Lags behind Europe by £40bn a Year’ Financial Times (16 November 2022) <www.ft.com/content/f752a1ad-4a23-408f-a549-4909974c6a2c> accessed 23 April 2024.
[21] ‘Why Is the NHS in Its Worst Ever Crisis?’, Financial Times (3 January 2023) <www.ft.com/content/b593116d-f948-4757-b2fa-c74adadc8b42> accessed 23 April 2024.
[22] Esme Kirk-Wade, Rachael Harker and Sonja Stiebahl, ‘NHS Key Statistics: England July 2024’ (House of Commons Library, July 2024) <https://researchbriefings.files.parliament.uk/documents/CBP-7281/CBP-7281.pdf> accessed 2 August 2024.
[23] Our Future Health, ‘The UK’s Largest Ever Health Research Programme’ (n 2).
[24] UK Research and Innovation, ‘Accelerating Detection of Disease’ (n 1).
[25] Ben Armstrong, ‘Polygenic Score Pilot for Heart Disease Begins’ (Genomics Education Programme, 28 January 2022) <www.genomicseducation.hee.nhs.uk/blog/polygenic-score-pilot-for-heart-disease-begins> accessed 3 August 2024.
[26] Office for Life Sciences and others, ‘Genome UK’ (n 4).
[27] NHS England, ‘Accelerating Genomic Medicine in the NHS’ (n 5).
[28] Turner and others, ‘Pharmacogenomics’ (n 8).
[29] Genomics England, ‘UK to Become World Number One in DNA Testing with Plan to Revolutionise Fight against Cancer and Rare Diseases’ (1 August 2014) <www.genomicsengland.co.uk/news/uk-becomes-number-one-in-dna-testing> accessed 29 April 2024.
[30] Department for Science, Innovation and Technology, DHSC and Office for Life Sciences, ‘Life Sciences Competitiveness Indicators 2023’ (July 2023) <www.gov.uk/government/publications/life-sciences-sector-data-2023/life-sciences-competitiveness-indicators-2023> accessed 29 April 2024.
[31] Orli G Bahcall, ‘UK Biobank: A New Era in Genomic Medicine’ (2018) 19(12), 737 Nature Reviews Genetics <https://doi.org/10.1038/s41576-018-0065-3> accessed 2 August 2024.
[32] Barclays and Eagle Labs, ‘Understanding the UK’s High-Growth Artificial Intelligence Companies’ (November 2023) <https://labs.uk.barclays/learning-and-insights/news-and-insights/thought-leadership/understanding-the-uks-high-growth-artificial-intelligence-companies> accessed 29 April 2024.
[33] Office for Life Sciences and others, ‘Genome UK: 2021 to 2022 Implementation Plan’ (May 2021) <www.gov.uk/government/publications/genome-uk-2021-to-2022-implementation-plan/genome-uk-2021-to-2022-implementation-plan> accessed 29 April 2024.
[34] Matt Davies, ‘A Lost Decade? The UK’s Industrial Approach to AI’ (AI Now Institute, 12 March 2024) <https://ainowinstitute.org/publication/a-lost-decade-the-uks-industrial-approach-to-ai> accessed 3 August 2024.
[35] Office for Life Sciences and others, ‘Genome UK’ (n 4).
[36] ‘Dr. Raghib Ali: “We Can Change the Whole Paradigm of Healthcare”’ (Our Future Health, 27 July 2022) <https://ourfuturehealth.org.uk/news/dr-raghib-ali-we-can-change-the-whole-paradigm-of-healthcare> accessed 21 April 2024.
[37] Nuffield Council on Bioethics, ‘Towards a Gold Standard of Ethics across Genomic Healthcare and Research: Where Are We?’ (2024) <www.nuffieldbioethics.org/assets/pdfs/NCOB-Genomics-Mapping-Report-Final-Web-PDF-Jan-2024.pdf> accessed 2 August 2024.
[38] Nuffield Council on Bioethics, ‘Resource Bank’ (2024) <www.nuffieldbioethics.org/assets/pdfs/NCOB-Resource-Bank-June-2024-FINAL-PUBLISHED.pdf> accessed 2 August 2024.
[39] Sowmiya Moorthie, ‘Application of Polygenic Scores in Healthcare’ (PHG Foundation, October 2023) <www.phgfoundation.org/wp-content/uploads/2024/04/2-Application-of-polygenic-scores-in-healthcare.pdf> accessed 2 August 2024.
[40] This issue is compounded by the fact that the common traits that polygenic scores are often used to predict are typically multidimensional, determined by many factors other than genomics, such as environmental factors.
[41] Genomics Education Programme, ‘Four Types of Genomic Testing Explained’ (Genomics Education Programme, 20 May 2019) <www.genomicseducation.hee.nhs.uk/blog/four-types-of-genomic-testing-explained> accessed 16 April 2024.
[42] In the UK, 2.4 million children and adults are estimated to be living with a genetic condition, roughly 3.6 per cent of the UK population of approximately 67 million. ‘About Genetic Disorders UK’ (Gene People), <www.genepeople.org.uk/about-us> accessed 6 December 2023.
[43] Peter M Visscher and others, ‘Discovery and Implications of Polygenicity of Common Diseases’ (2021) 373(6562), 1468 Science <https://doi.org/10.1126/science.abi8206> accessed 2 August 2024.
[44] Sowmiya Moorthie and others, ‘Polygenic Scores, Risk and Cardiovascular Disease’ (PHG Foundation, August 2019).
[45] Moorthie, ‘Application of Polygenic Scores in Healthcare’ (n 39).
[46] Chantal Babb de Villiers and others, ‘Polygenic Scores for Cancer’ (PHG Foundation, September 2022) <www.phgfoundation.org/wp-content/uploads/2023/10/Polygenic-scores-for-cancer.pdf> accessed 2 August 2024.
[47] Sobia Raza, ‘Artificial Intelligence for Genomic Medicine’ (PHG Foundation, March 2020) <www.phgfoundation.org/wp-content/uploads/2024/02/Artifical-intelligence-for-genomic-medicine.pdf> accessed 2 August 2024.
[48] Farmer, DNA.I. (n 7).
[49] DHSC, ‘Genome UK: 2022 to 2025 Implementation Plan for England’ (December 2022) <www.gov.uk/government/publications/genome-uk-2022-to-2025-implementation-plan-for-england/genome-uk-2022-to-2025-implementation-plan-for-england> accessed 23 May 2024.
[50] NHS England, ‘Accelerating Genomic Medicine in the NHS’ (n 5).
[51] Ben Armstrong, ‘NHS Launches New Polygenic Scores Trial for Heart Disease’ (Genomics Education Programme, 29 April 2021) <www.genomicseducation.hee.nhs.uk/blog/nhs-launches-new-polygenic-scores-trial-for-heart-disease> accessed 3 August 2024.
[52] ‘Polygenic risk scores performed poorly in population screening, individual risk prediction, and population risk stratification. Strong claims about the effect of polygenic risk scores on healthcare seem to be disproportionate to their performance.’ Aroon D Hingorani and others, ‘Performance of Polygenic Risk Scores in Screening, Prediction, and Risk Stratification: Secondary Analysis of Data in the Polygenic Score Catalog’ (2023) 2(1) BMJ Medicine <https://doi.org/10.1136/bmjmed-2023-000554> accessed 2 August 2024.
[53] ‘For majority of common diseases and quantitative traits, PGS currently have relatively low overall prediction accuracy across individuals in the general population.’ Ying Ma and Xiang Zhou, ‘Genetic Prediction of Complex Traits with Polygenic Scores: A Statistical Review’ (2021) 37(11), 995 Trends in Genetics <https://doi.org/10.1016/j.tig.2021.06.004> accessed 2 August 2024.
[54] ‘Genomics beyond Health’ (Government Office for Science, January 2022) <www.gov.uk/government/publications/genomics-beyond-health> accessed 17 April 2024.
[55] ‘[T]he accuracy of PGS is expected to improve along with increasing GWAS sample size, availability of new genomic information from omics studies, as well as the development of advanced PGS methods.’ Ma and Zhou, ‘Genetic Prediction’ (n 53).
[56] ‘[T]he number of variants tested has increased 20-fold; from ~500,000 variants in the early days to nearly 10 million variants in the latest GWASs.’ Ruth JF Loos, ‘15 Years of Genome-Wide Association Studies and No Signs of Slowing Down’ (2020) 11(1), 5900 Nature Communications <https://doi.org/10.1038/s41467-020-19653-5> accessed 2 August 2024.
[57] The cost to sequence a whole human genome decreased by 97.7 per cent between 2010 and 2022; ‘Genomics beyond Health’ (n 54).
[58] Claudia Caudai and others, ‘AI Applications in Functional Genomics’ (2021) 19, 5762 Computational and Structural Biotechnology Journal <https://doi.org/10.1016/j.csbj.2021.10.009> accessed 2 August 2024.
[59] Harini Suresh and John Guttag, ‘A Framework for Understanding Sources of Harm throughout the Machine Learning Life Cycle’ in EAAMO ’21: Proceedings of the 1st ACM Conference on Equity and Access in Algorithms, Mechanisms, and Optimization (October 2021) <https://doi.org/10.1145/3465416.3483305> accessed 2 August 2024.
[60] Irene Y Chen, Peter Szolovits and Marzyeh Ghassemi, ‘Can AI Help Reduce Disparities in General Medical and Mental Health Care?’ (2019) 21(2), E167 AMA Journal of Ethics <https://doi.org/10.1001/amajethics.2019.167> accessed 2 August 2024.
[61] Yi Ding and others, ‘Polygenic Scoring Accuracy Varies across the Genetic Ancestry Continuum’ 2023) 618(7966), 774 Nature <https://doi.org/10.1038/s41586-023-06079-4> accessed 2 August 2024.
[62] Ying Wang and others, ‘Challenges and Opportunities for Developing More Generalizable Polygenic Risk Scores’ (2022) 5, 293 Annual Review of Biomedical Data Science <https://doi.org/10.1146/annurev-biodatasci-111721-074830> accessed 2 August 2024.
[63] Melinda C Mills and Charles Rahal, ‘A Scientometric Review of Genome-Wide Association Studies’ (2019) 2(1), 9 Communications Biology <https://doi.org/10.1038/s42003-018-0261-x> accessed 2 August 2024.
[64] Elliott Williams, ‘Mind the Gap: Five Initiatives to Boost Genomic Data Diversity’ (Genomics Education Programme, 22 March 2024) <www.genomicseducation.hee.nhs.uk/blog/mind-the-gap-five-initiatives-to-boost-genomic-data-diversity> accessed 3 August 2024.
[65] Carol Brayne and Terrie E Moffitt, ‘The Limitations of Large-Scale Volunteer Databases to Address Inequalities and Global Challenges in Health and Aging’ (2022) 2(9), 775 Nature Aging <https://doi.org/10.1038/s43587-022-00277-x> accessed 2 August 2024.
[66] Anneke Lucassen, ‘Utility of Polygenic Scores in Healthcare’ (National Screening, February 2024) <https://nationalscreening.blog.gov.uk/wp-content/uploads/sites/254/2024/02/Utility-of-polygenic-scores-in-healthcare-presentation-6-2-24_.pdf> accessed 2 August 2024.
[67] Some statistical methods, such as penalised elastic net regression models, attempt to address this problem. See: Tanigawa Y and others, ‘Significant Sparse Polygenic Risk Scores across 813 Traits in UK Biobank’ (2022) 18 PLOS Genetics e1010105 <https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1010105> accessed 5 August 2024.
[68] Farmer, DNA.I. (n 7).
[69] Raza, ‘Artificial Intelligence’ (n 47).
[70] Tingting Guo and Xianran Li, ‘Machine Learning for Predicting Phenotype from Genotype and Environment’ (2023) 79, 102853 Current Opinion in Biotechnology <https://doi.org/10.1016/j.copbio.2022.102853> accessed 2 August 2024.
[71] Daniel Sik Wai Ho and others, ‘Machine Learning SNP Based Prediction for Precision Medicine’ 2019 10, 267 Frontiers in Genetics <https://doi.org/10.3389/fgene.2019.00267> accessed 2 August 2024.
[72] ‘Current [polygenic scoring] models typically build on combining genetics and electronic health record (EHR) features additively (i.e., a simple summation), leaving room for the development of more complete approaches, for example a deep neural network (DNN) that takes as input the different risk factors jointly to learn about the complex interplay between them.’ Marie-Christine Fritzsche and others, ‘Ethical Layering in AI-Driven Polygenic Risk Scores: New Complexities, New Challenges’ 2023 14 Frontiers in Genetics <https://doi.org/10.3389/fgene.2023.1098439> accessed 2 August 2024.
[73] Raza, ‘Artificial Intelligence’ (n 47).
[74] Turner and others, ‘Pharmacogenomics’ (n 8).
[75] For instance, an individual deciding to take a genomic test for a genetic disease may inadvertently reveal whether a close relative has that disease. In such cases, a person’s interest in understanding their health might conflict with their relative’s desire to not know about theirs. It also means that people who want to keep their genomic details private may struggle if their relatives are more willing to share the results of genomic tests.
[76] A notorious example of this is Target Mart’s ability to determine when customers were pregnant from patterns in data from loyalty cards; Brett Belock and others, Target Corporation: Predictive Analytics and Customer Privacy (Eugene D. Fanning Center for Business Communication, Mendoza College of Business, University of Notre Dame, 2013) <https://doi.org/10.4135/9781526403568> accessed 2 August 2024.
[77] ‘About Genetic Disorders UK’ (n 42).
[78] ‘Genomics beyond Health’ (n 54).
[79] Under the GDPR and UK data protection law, the term ‘sensitive data’ is used interchangeably with ‘special category personal data’. This includes personal data revealing racial or ethnic origin, political opinions, or religious or philosophical beliefs; trade union membership; genetic data, biometric data processed solely to identify a human being; health-related data; and data concerning a person’s sex life or sexual orientation.
[80] Luc Rocher, Julien M Hendrickx and Yves-Alexandre De Montjoye, ‘Estimating the Success of Re-Identifications in Incomplete Datasets using Generative Models’ 2019 10(1), 3069 Nature Communications <https://doi.org/10.1038/s41467-019-10933-3> accessed 2 August 2024.
[81] Ruichu Cai and others, ‘Deterministic Identification of Specific Individuals from GWAS Results’ (2015) 31(11), 1701 Bioinformatics <https://doi.org/10.1093/bioinformatics/btv018> accessed 2 August 2024.
[82] A good illustration of how these kinds of pressures might manifest can be found in ‘Innovation Should Support Societal Responsibility for Health’ (The BMJ, 30 December 2019) <https://blogs.bmj.com/bmj/2019/12/30/innovation-support-societal-responsibility-health> accessed 23 May 2024.
[83] Many (though by no means all) participants in our public engagement exercise expressed the view that individuals have a degree of responsibility or moral obligation to share their genomic data to improve the quality of healthcare for everybody.
[84] Atiya Kamal, Ava Hodson and Julia M Pearce, ‘A Rapid Systematic Review of Factors influencing COVID-19 Vaccination Uptake in Minority Ethnic Groups in the UK’ (2021) 9(10), 1121 Vaccines.
[85] One reason is that when money is tight, it can be very difficult to justify not choosing the cheapest option, even if it comes with significant non-monetary costs. Another reason is that for those who fail to disclose genomic data, health insurance may well be not just more expensive than the discounted rates available to those who do share their genomic data but also more expensive than previous, non-genomic rates of insurance. This is because failure to disclose genomic information could be interpreted by the insurer as evidence that the customer has a poor disease risk score that they want to hide.
[86] ICO, ‘What Is Special Category Data?’ (9 April 2024) <https://ico.org.uk/for-organisations/uk-gdpr-guidance-and-resources/lawful-basis/special-category-data/what-is-special-category-data> accessed 10 April 2024.
[87] Beatrice Kaiser and others, ‘A Proposal for an Inclusive Working Definition of Genetic Discrimination to Promote a More Coherent Debate’ (2024) 56, 1339 Nature Genetics <https://doi.org/10.1038/s41588-024-01786-8> accessed 2 August 2024.
[88] Ada Lovelace Institute, ‘Tackling Health and Social Inequalities in Data-Driven Systems’ <www.adalovelaceinstitute.org/project/health-social-inequalities-data-driven-systems> accessed 8 May 2024.
[89] D Clark, ‘Inequality in the UK: Statistics and Facts’ (Statista, December 2023) <www.statista.com/topics/8436/inequality-in-the-uk> accessed 7 May 2024.
[90] David Finch and Adam Tinson, ‘The Continuing Impact of COVID-19 on Health and Inequalities’ (The Health Foundation, August 2022) <www.health.org.uk/publications/long-reads/the-continuing-impact-of-covid-19-on-health-and-inequalities> accessed 3 August 2024.
[91] Scientific Advisory Group for Emergencies, ‘Drivers of the Higher COVID-19 Incidence, Morbidity and Mortality among Minority Ethnic Groups, 23 September 2020’ (May 2022) <www.gov.uk/government/publications/drivers-of-the-higher-covid-19-incidence-morbidity-and-mortality-among-minority-ethnic-groups-23-september-2020/drivers-of-the-higher-covid-19-incidence-morbidity-and-mortality-among-minority-ethnic-groups-23-september-2020–2> accessed 7 May 2024.
[92] ‘Insurers are economically motivated to accurately classify risk or jeopardize losing consumers to their competition, leaving risk classification models susceptible to an arms-race mentality between insurers … Genomic data feeds directly into the frenzy of the risk classification arms race, foreshadowing a potential transformation of how insurers classify risk.’ Anya ER Prince, ‘Insurance Risk Classification in an Era of Genomics: Is a Rational Discrimination Policy Rational?’ (2017) 96(3), 624 Nebraska Law Review.
[93] Nina Jais and others, Insuring Shared Value: How Insurers Gain Competitive Advantage by Better Addressing Society’s Needs (FSG, 2017) <www.fsg.org/resource/insuring-shared-value> accessed 2 August 2024.
[94] ‘Vitality’s Shared-Value Insurance Encourages Behavior-Change to Make People Healthier’ (Vitality) <https://vitality.international/about-vitality/shared-value-insurance.html> accessed 26 July 2024.
[95] ‘Car Insurance from Insurethebox’ (Insurethebox) <www.insurethebox.com> accessed 26 July 2024.
[96] Lamiece Hassan and others, ‘A Deliberative Study of Public Attitudes towards Sharing Genomic Data within NHS Genomic Medicine Services in England’ (2020) 29(7), 702 Public Understanding of Science <https://doi.org/10.1177/0963662520942132> accessed 2 August 2024.
[97] Anya ER Prince, ‘Comparative Perspectives: Regulating Insurer Use of Genetic Information’ 2019) 27(3), 340 European Journal of Human Genetics <https://doi.org/10.1038/s41431-018-0293-1> accessed 2 August 2024.
[98] ‘Australians Shun Genetic Testing, Fearing Impact on Life Insurance’ ABC (30 June 2023) <www.abc.net.au/news/2023-06-30/australians-shun-genetic-testing-fearing-impact-on-insurance/102546976> accessed 3 August 2024.
[99] DHSC, ‘Results of the Code on Genetic Testing and Insurance Call for Evidence’ (April 2024) <www.gov.uk/government/calls-for-evidence/code-on-genetic-testing-and-insurance-call-for-evidence/outcome/results-of-the-code-on-genetic-testing-and-insurance-call-for-evidence> accessed 7 May 2024.
[100] Turo-Kimmo Lehtonen and Jyri Liukko, ‘The Forms and Limits of Insurance Solidarity’ 2011) 103(S1), 33 Journal of Business Ethics <https://doi.org/10.1007/s10551-012-1221-x> accessed 2 August 2024.
[101] Jyri Liukko, ‘Genetic Discrimination, Insurance, and Solidarity: An Analysis of the Argumentation for Fair Risk Classification’ (2010) 29(4), 457 New Genetics and Society <https://doi.org/10.1080/14636778.2010.528197> accessed 2 August 2024.
[102] ‘Insurance is a mechanism of distributive justice and mutual aid whereby individuals come together to increase access to social goods and economic security through redistribution from the lucky to the unlucky – from those who pay insurance premiums, but never face a loss, to those who experience sickness, death, or other insured harm and must file claims.’ Prince, ‘Insurance Risk Classification’ (n 93).
[103] The idea that private insurance should serve as a redistributive mechanism is not subscribed to universally. However, arguments for a degree of solidarity in insurance are likely to be more persuasive (and more accepted) when the object of insurance is a basic human necessity such as healthcare, and where the factors determining people’s differing levels of risk are matters of ‘brute luck’ rather than active choices. Both of these criteria apply to the use of AIGHP by health insurers.
[104] Richard Ashcroft, ‘Should Genetic Information Be Disclosed to Insurers? No’ (2007) 334(7605), 1197 BMJ <https://doi.org/10.1136/bmj.39216.425231.AD> accessed 2 August 2024.
[105] Jais and others, Insuring Shared Value (n 94).
[106] J Morley, ‘On Designing an Algorithmically Enhanced NHS: Towards a Conceptual Model for the Successful Implementation of Algorithmic Clinical Decision Support Software in the National Health Service’ (Oxford University Research Archive, 2023) <https://ora.ox.ac.uk/objects/uuid:0f58b2c4-ffa0-412d-afc3-aedc2eaf46d4> accessed 2 August 2024.
[107] Giovanni Rubeis, ‘Liquid Health: Medicine in the Age of Surveillance Capitalism’ (2023) 322, 115810 Social Science & Medicine <https://doi.org/10.1016/j.socscimed.2023.115810> accessed 2 August 2024.
[108] Jeanette Kennett, ‘Undeserving Patients’ in B Davies and others (eds) Responsibility and Healthcare (Oxford University Press, 2024), 61.
[109] ‘Rationing Healthcare to Smokers and Obese?’ BBC News (14 November 2017) <www.bbc.co.uk/news/av/uk-politics-41988545> accessed 8 February 2024.
[110] Bianca Sjoerdstra, ‘Dealing with Vendor Lock-In’ (BS thesis, University of Twente, 2016) <http://essay.utwente.nl/70153> accessed 2 August 2024.
[111] Lindsay Clark, ‘Vendor Lock-in Hurts UK Govt Ability to Negotiate Spending’ The Register (April 2024) <www.theregister.com/2024/04/04/uk_cddo_admits_cloud_spending_lock_issues_exclusive> accessed 3 August 2024.
[112] Nicola Byrne, ‘The NHS Federated Data Platform: The Importance of Building Bridges with the Public’ (National Data Guardian, August 2023) <www.gov.uk/government/news/the-nhs-federated-data-platform-the-importance-of-building-bridges-with-the-public> accessed 3 August 2024.
[113] Oscar Williams, ‘NHS Deal with Palantir Raises Fears of Vendor Lock-in’ (Privacy International, 27 April 2020) <http://privacyinternational.org/examples/3814/nhs-deal-palantir-raises-fears-vendor-lock> accessed 3 August 2024.
[114] Alex Hern, ‘Royal Free Breached UK Data Law in 1.6m Patient Deal with Google’s DeepMind’ Guardian (3 July 2017) <www.theguardian.com/technology/2017/jul/03/google-deepmind-16m-patient-royal-free-deal-data-protection-act> accessed 3 August 2024.
[115] Chris Smyth, ‘Tony Blair and William Hague: Sell NHS Data to Fund Medical Advances’ The Times (25 January 2024) <www.thetimes.co.uk/article/tony-blair-and-william-hague-sell-nhs-data-to-fund-medical-advances-fz27bmb98> accessed 3 August 2024.
[116] Anna Middleton and others, ‘Global Public Perceptions of Genomic Data Sharing: What Shapes the Willingness to Donate DNA and Health Data?’ (2020) 107(4), 742 American Journal of Human Genetics <https://doi.org/10.1016/j.ajhg.2020.08.023> accessed 2 august 2024.
[117] Hassan and others, ‘A Deliberative Study’ (n 97).
[118] Nick Triggle, ‘Care.data: How Did It Go So Wrong?’ BBC News (19 February 2014) <www.bbc.com/news/health-26259101> accessed 3 August 2024.
[119] Hern, ‘Royal Free Breached UK Data Law’ (n 115).
[120] Julia Powles and Hal Hodson, ‘Google DeepMind and Healthcare in an Age of Algorithms’ (2017) 7(4), 351 Health and Technology <https://doi.org/10.1007/s12553-017-0179-1> accessed 2 August 2024.
[121] Chaminda Jayanetti, ‘NHS Data Grab on Hold as Millions Opt Out’ Observer (22 August 2021) <www.theguardian.com/society/2021/aug/22/nhs-data-grab-on-hold-as-millions-opt-out> accessed 3 August 2024.
[122] Denis Campbell, ‘NHS Data Platform May Be Undermined by Lack of Public Trust, Warn Campaigners’ Guardian (21 November 2023) <www.theguardian.com/society/2023/nov/21/nhs-data-platform-may-be-undermined-by-lack-of-public-trust-warn-campaigners> accessed 3 August 2024.
[123] Simon Chesterman, ‘Through a Glass, Darkly: Artificial Intelligence and the Problem of Opacity’ (2021) 69(2), 271 The American Journal of Comparative Law.
[124] Helen Smith, ‘Clinical AI: Opacity, Accountability, Responsibility and Liability’ (2021) 36(2), 535 AI & Society <https://doi.org/10.1007/s00146-020-01019-6> accessed 2 August 2024.
[125] ‘[Healthcare] providers may develop too much reliance or trust on a CDSS [Clinical Decision Support System] for a specific task. This could be compared to using a calculator for mathematical operations over a long period of time, and then having poorer mental math skills. It is potentially problematic as the user has less independence and will be less equipped for that task should they switch to an environment without the CDSS.’ Reed T Sutton and others, ‘An Overview of Clinical Decision Support Systems: Benefits, Risks, and Strategies for Success’ (2020) 3(1), 1 NPJ Digital Medicine <https://doi.org/10.1038/s41746-020-0221-y> accessed 2 August 2024.
[126] Morley, ‘On Designing an Algorithmically Enhanced NHS’ (n 107).
[127] ‘Relying on ACDSS [Adaptive CDSS] may mean that where once patient-led reports of the haptic “sensations” they felt in their bodies would have been considered valid and relevant knowledge of the body, now the only knowledge considered “valid” is that which is “quantifiable and observable” (i.e., positivistic) (Chin-Yee & Upshur, 2019). Humanistic knowledge (such as a person’s lived experience (Deeny & Steventon, 2015) or their subjective feeling 114 of wellbeing (Molnár-Gábor, 2020b) or common sense knowledge (R. D. Schwartz, 1989), may be deemed irrelevant and ignorable.’ Ibid.
[128] Benjamin Chin‐Yee and Ross Upshur, ‘Clinical Judgement in the Era of Big Data and Predictive Analytics’ (2018) 24(3), 638 Journal of Evaluation in Clinical Practice <https://doi.org/10.1111/jep.12852> accessed 2 August 2024.
[129] Denis Campbell, ‘Private Healthcare Could Become “a New Normal” as NHS Grows Weaker’ Guardian (8 March 2024) <www.theguardian.com/society/2024/mar/08/private-healthcare-could-become-a-new-normal-as-nhs-grows-weaker> accessed 3 August 2024.
[130] Maria Davies, ‘Healthcode Data Indicates Record Insured Activity in 2023’ LaingBuisson News (21 February 2024) <www.laingbuissonnews.com/healthcare-markets-content/healthcode-data-indicates-record-insured-activity-in-2023> accessed 3 August 2024.
[131] ‘Patients Drift towards Paying for Hospital Care out of Their Own Pocket across All Four UK Countries’ (Nuffield Trust, 16 May 2024) <www.nuffieldtrust.org.uk/news-item/patients-drift-towards-paying-for-hospital-care-out-of-their-own-pocket-across-all-four-uk-countries> accessed 18 June 2024.
[132] Office for Life Sciences and others, ‘Genome UK’ (n 4).
[133] ‘Dr. Raghib Ali’ (n 36).
[134] Amit Sud and others, ‘Realistic Expectations Are Key to Realising the Benefits of Polygenic Scores’ (2023) e073149 BMJ <https://doi.org/10.1136/bmj-2022-073149> accessed 2 August 2024.
[135] Genomics PLC, ‘Polygenic Risk Scores: White Paper’ (March 2019) <https://26790458.fs1.hubspotusercontent-eu1.net/hubfs/26790458/Genomicsplc_August2023/Pdf/Genomics-plc-PRS-details_White-Paper-April-2019.pdf> accessed 2 August 2024.
[136] ‘[M]ost people who develop disease will not have a high polygenic score.’ Sud and others, ‘Realistic Expectations’ (n 135).
[137] Ole‐Jorgen Skog, ‘The Prevention Paradox Revisited’ (1999) 95(5), 751 Addiction <https://doi.org/10.1046/j.1360-0443.1999.94575113.x> accessed 2 August 2024.
[138] Stephen John, ‘Why the Prevention Paradox Is a Paradox, and Why We Should Solve It: A Philosophical View’ (2011) 53(4–5), 250 Preventive Medicine <https://doi.org/10.1016/j.ypmed.2011.07.006> accessed 2 August 2024.
[139] Nilanjan Chatterjee, Jianxin Shi and Montserrat García-Closas, ‘Developing and Evaluating Polygenic Risk Prediction Models for Stratified Disease Prevention’ (2016) 17(7), 392 Nature Reviews: Genetics <https://doi.org/10.1038/nrg.2016.27> accessed 3 August 2024.
[140] ‘Polygenic Risk Predictions: Health Revolution or Going Round in Circles?’ (Genewatch, September 2023) <www.genewatch.org/uploads/f03c6d66a9b354535738483c1c3d49e4/gw-prs-briefing-fin.pdf> accessed 2 August 2024.
[141] H Gilbert Welch, ‘Cancer Screening: The Good, the Bad, and the Ugly’ (2022) 157(1), 467 JAMA Surgery <https://doi.org/10.1001/jamasurg.2022.0669> accessed 2 August 2024.
[142] ‘Polygenic Risk Predictions’ (n 141).
[143] ‘Dr. Raghib Ali’ (n 36).
[144] ‘Meta-analysis revealed no significant effects of communicating DNA based risk estimates on smoking cessation (odds ratio 0.92, 95% confidence interval 0.63 to 1.35, P=0.67), diet (standardised mean difference 0.12, 95% confidence interval −0.00 to 0.24, P=0.05), or physical activity (standardised mean difference −0.03, 95% confidence interval −0.13 to 0.08, P=0.62). There were also no effects on any other behaviours (alcohol use, medication use, sun protection behaviours, and attendance at screening or behavioural support programmes) or on motivation to change behaviour.’ Gareth J Hollands and others, ‘The Impact of Communicating Genetic Risks of Disease on Risk-Reducing Health Behaviour: Systematic Review with Meta-Analysis’ (2016) i1102 BMJ <https://doi.org/10.1136/bmj.i1102> accessed 2 August 2024.
[145] ‘[The effects of nudging] on more complex, continuing behaviours such as self-management of chronic conditions remain unclear. In addition, long term studies are still lacking for many nudging techniques, and new evidence suggests that some nudges may not be as effective as originally thought when implemented outside experimental settings.’ Thomas Rouyard and others, ‘Boosting Healthier Choices’ (2022) e064225 BMJ <https://doi.org/10.1136/bmj-2021-064225> accessed 2 August 2024.
[146] For instance, being informed that your diet is unhealthy and receiving nudges to eat better have limited value if you are unable to afford healthier foods, lack the time to cook or live in an area with few shops in which to buy fresh ingredients.
[147] Barbara Prainsack, ‘The Value of Healthcare Data: To Nudge, or Not?’ (2020) 41(5), 547 Policy Studies <https://doi.org/10.1080/01442872.2020.1723517> accessed 2 August 2024.
[148] ‘People who do not have “high risk” polygenic scores might be less likely to seek medical attention for concerning symptoms, or their clinicians might be less inclined to investigate.’ Sud and others, ‘Realistic Expectations’ (n 130).
[149] European Parliament and Council of the European Union, ‘Regulation (EU) 2016/679 of the European Parliament and of the Council of 27 April 2016 on the Protection of Natural Persons with Regard to the Processing of Personal Data and on the Free Movement of Such Data (United Kingdom General Data Protection Regulation)(Text with EEA Relevance)’ <www.legislation.gov.uk/eur/2016/679/contents> accessed 10 April 2024.
[150] Colin Mitchell and others, ‘The GDPR and Genomic Data’ (PHG Foundation, April 2020) <www.phgfoundation.org/publications/reports/the-gdpr-and-genomic-data> accessed 3 August 2024.
[151] ICO, ‘What Is Personal Data?’ (19 May 2023) <https://ico.org.uk/for-organisations/uk-gdpr-guidance-and-resources/personal-information-what-is-it/what-is-personal-data/what-is-personal-data> accessed 2 May 2024.
[152] The Prime Minister’s Office, ‘The King’s Speech 2024’ (July 2024) <https://assets.publishing.service.gov.uk/media/6697f5c10808eaf43b50d18e/The_King_s_Speech_2024_background_briefing_notes.pdf> accessed 2 August 2024.
[153] The DPDI Bill specifies that where a living individual ‘may be identified directly or indirectly’, that data is personal data both where the controller or processor could identify the information ‘by reasonable means’ and ‘where the controller or processor knows, or ought reasonably to know, that another person is likely to obtain the information because of the processing and could identify an individual by reasonable means’. Here, ‘by reasonable means’ is a test that is supposed to take account of ‘the time, effort and cost to identify an individual from the information’ and ‘the technology and other resources available’. John Woodhouse, ‘Research Briefing on the Data Protection and Digital Information Bill’ (House of Commons Library, August 2022) <https://researchbriefings.files.parliament.uk/documents/CBP-9606/CBP-9606.pdf> accessed 2 August 2024.
[154] It also requires an assessment of what it takes for it to be likely that a given agent might obtain the data and what means would be reasonable for them.
[155] Mitchell and others, ‘The GDPR and Genomic Data’ (n 151).
[156] Because the development of AIGHP systems involves identifying correlations between genotype and phenotype, good-quality phenotype data is indispensable for AIGHP research. In addition to being necessary for the development of AIGHP systems, accurate predictions about given individuals using AIGHP may well depend on combining both genomic and phenotype data (combined and integrated risk scores), making phenotype data important for the deployment as well as for the development of AIGHP systems.
[157] European Parliament and Council of the European Union, ‘Regulation (EU) 2016/679′, Article 4(15) (n 150).
[158] Sachin H Jain and others, ‘The Digital Phenotype’ (2015) 33(5), 462 Nature Biotechnology <https://doi.org/10.1038/nbt.3223> accessed 2 August 2024.
[159] Hunton Andrews Kurth LLP, ‘Article 29 Working Party Clarifies Scope of Health Data Processed by Lifestyle and Wellbeing Apps’ (Privacy & Information Security Law Blog, 9 February 2015) <www.huntonprivacyblog.com/2015/02/09/article-29-working-party-clarifies-scope-health-data-processed-lifestyle-wellbeing-apps> accessed 10 April 2024.
[160] In its analysis of the working party’s guidance, law firm Covington and Burling put this point more mildly: ‘The Working Party’s criteria for the definition establish a rather low threshold for information in apps and devices to qualify as health data.’ Covington & Burling LLP, ‘Article 29 Working Party Clarifies Scope of Health Data in Apps and Devices’ (National Law Review, February 2015) <www.natlawreview.com/article/article-29-working-party-clarifies-scope-health-data-apps-and-devices> accessed 3 August 2024.
[161] ICO, ‘What Is Special Category Data?’ (n 87).
[162] Ibid.
[163] Direct-to-consumer genetic testing refers to genetic and genomic tests offered directly to members of the public by private companies such as ancestry.com and, historically, 23andMe. There are concerns that some direct-to-consumer tests provide misleading or inaccurate results to users.
[164] Juan Ramón Robles, ‘Compatibility Test: Can I Process Lawfully Collected Personal Data for a New Purpose?’ (Hogan Lovells, 17 May 2021) <www.engage.hoganlovells.com/knowledgeservices/news/compatibility-test-can-i-process-lawfully-collected-personal-data-for-a-new-purpose_1> accessed 7 May 2024.
[165] ‘[R]ather than imposing a requirement of compatibility, the legislator chose a double negation: it prohibited incompatibility. By providing that any further processing is authorised as long as it is not incompatible (and if the requirements of lawfulness are simultaneously also fulfilled), it would appear that the legislators intended to give some flexibility with regard to further use … The fact that further processing is for a different purpose does not necessarily mean that it is automatically incompatible.’ Article 29 Data Protection Working Party, ‘Opinion 03/2013 on Purpose Limitation’ (April 2013) <https://ec.europa.eu/justice/article-29/documentation/opinion-recommendation/files/2013/wp203_en.pdf> accessed 2 August 2024.
[166] The new Article 8A(3) states that ‘processing of personal data for a new purpose is to be treated in a manner compatible with the original purpose where … the processing is carried out in accordance with Article 84B – for the purposes of scientific research or historical research.’ UK Parliament, ‘Data Protection and Digital Information Bill’ <https://bills.parliament.uk/bills/3430> accessed 11 April 2024.
[167] Prior to the Act, Article 13(3) of the UK GDPR stated that: ‘Where the controller intends to further process the personal data for a purpose other than that for which the personal data were collected, the controller shall provide the data subject prior to that further processing with information on that other purpose and with any relevant further information as referred to in paragraph 2’. A new Article 13(5) has now been added which states that this obligation does not apply under various circumstances, including where the further processing is conducted ‘for (and only for) the purposes of scientific or historical research, the purposes of archiving in the public interest or statistical purposes’.
[168] AI Law Consultancy, commissioned legal analysis of the Data Protection and Digital Information Bill.
[169] It could also be argued that the all-or-nothing nature of sharing genomic data for research weakens the force of a subject’s having consented to processing: when a particular choice is bundled up with several others, it is hard to disentangle an actor’s true preferences. It might also be argued that the practical difficulties associated with predicting the uses to which a subject’s data might be put undermine the transparency of data sharing and processing.
[170] Office for Life Sciences and others, ‘Genome UK’ (n 4).
[171] ‘Equality Act 2010’ <www.legislation.gov.uk/ukpga/2010/15/contents> accessed 2 August 2024.
[172] Specifically, any changes to the law would need to avoid situations in which entities are unable to engage in forms of discrimination currently considered normal and legal because they could be argued to be instances of genomic discrimination. For instance, a potential employer might want to discriminate against a potential employee on the grounds of a particular phenotypic trait (e.g., that person’s performance on an aptitude test). Because performance on some kinds of aptitude tests could be argued to have a genomic basis, that employer might be open to the charge of genomic discrimination under a version of equalities law that considered genotype to be a protected characteristic.
[173] Equality Act 2010 (n 172).
[174] For a fuller picture of legislation on genetic and genomic discrimination around the world, see Genetic Discrimination Observatory, ‘A Geographical Overview of Approaches adopted around the World to Prevent Genetic Discrimination’ <https://gdo.global/en/gdo-map-approaches> accessed 3 August 2024.
[175] The code replaces a previous moratorium on genetic testing by insurers. The code states that insurers agree not to use the results of predictive genetic tests to inform insurance coverage decisions, except in very limited circumstances.
[176] DHSC, ‘Code on Genetic Testing and Insurance: 3-Year Review 2022’ (December 2022) <www.gov.uk/government/publications/code-on-genetic-testing-and-insurance-3-year-review-2022/code-on-genetic-testing-and-insurance-3-year-review-2022> accessed 5 June 2024.
[177] DHSC, ‘Results of the Code on Genetic Testing and Insurance Call for Evidence’ (n 100).
[178] Carlota Perez, Technological Revolutions and Financial Capital (Edward Elgar Publishing, 2003).
[179] Mariana Mazzucato, ‘From Market Fixing to Market-Creating: A New Framework for Economic Policy’ (SSRN Electronic Journal, 2015) https://doi.org/10.2139/ssrn.2744593> accessed 2 August 2024.
[180] In the NHS in England, Integrated Systems are local partnerships that bring health and care organisations together to develop shared plans and joined-up services.
[181] As we noted earlier in this report, however, the evidence is mixed concerning the extent to which knowledge of genomic disease risk affects behaviour.
[182] Tony Blair, ‘A Credible Plan to Transform Care Is the Best Birthday Present We Could Give the NHS’, Telegraph (6 July 2023) <www.telegraph.co.uk/politics/2023/07/06/tony-blair-nhs-anniversary-plan-transform-care> accessed 3 August 2024.
[183] ‘Fit for the Future: How a Healthy Population Will Unlock a Stronger Britain’ (Tony Blair Institute for Global Change, March 2023) <www.institute.global/insights/public-services/fit-future-how-healthy-population-will-unlock-stronger-britain> accessed 3 August 2024.
[184] Ibid.
[185] Samantha Field, ‘Public Health Gets Personal: The Case for an AI-Driven Personalised Prevention Platform’ Prospect (April 2024) <www.prospectmagazine.co.uk/sponsored/65662/public-health-gets-personal-the-case-for-an-ai-driven-personalised-prevention-platform> accessed 3 August 2024.
[186] Blair, ‘A Credible Plan (n 183).
[187] On one hand, the maximalist vision is one in which a person’s experience of healthcare (and what is offered to and expected of them) is more likely to be coloured by their genotype. On the other, there is a prima facie appealing universality to the maximalist vision’s insistence on providing access to AIGHP insight to every member of the population, rather than a few who it is deemed worthwhile for.
[188] DHSC, ‘Code on Genetic Testing and Insurance’ (n 177).
[189] The problem that the Government and the insurance industry is worried about arises not because of an asymmetry of information between insurance providers and customers but because individual-level knowledge of risk sits uneasily with voluntary risk pooling. In response to such an eventuality, the only real solution is to force people to engage in the insurance market through compulsory insurance, or to move away from an insurance-based model of healthcare.
[190] Amare Health, ‘The Number of People Using Private Healthcare Is on the Rise’ <https://amarehealth.co.uk/the-number-of-people-using-private-healthcare-is-on-the-rise> accessed 18 June 2024.
[191] Campbell, ‘Private Healthcare Could Become “a New Normal”’ (n 130).
[192] The prevalence of this view (that AIGHP may be better than the alternative) suggests many participants may think that given the current realities and the experience of the past decade of cuts to public services, the adoption of new technologies may be the only way of addressing current problems with healthcare in the UK.
Related content
DNA.I.
Early findings and emerging questions on the use of AI in genomics